The Evolutionary Basis of Bioinformatics An Introduction to
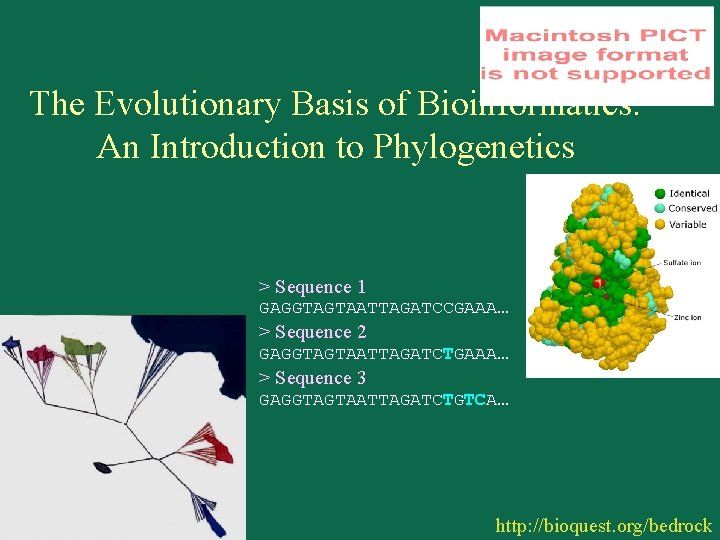
The Evolutionary Basis of Bioinformatics: An Introduction to Phylogenetics > Sequence 1 GAGGTAGTAATTAGATCCGAAA… > Sequence 2 GAGGTAGTAATTAGATCTGAAA… > Sequence 3 GAGGTAGTAATTAGATCTGTCA… http: //bioquest. org/bedrock
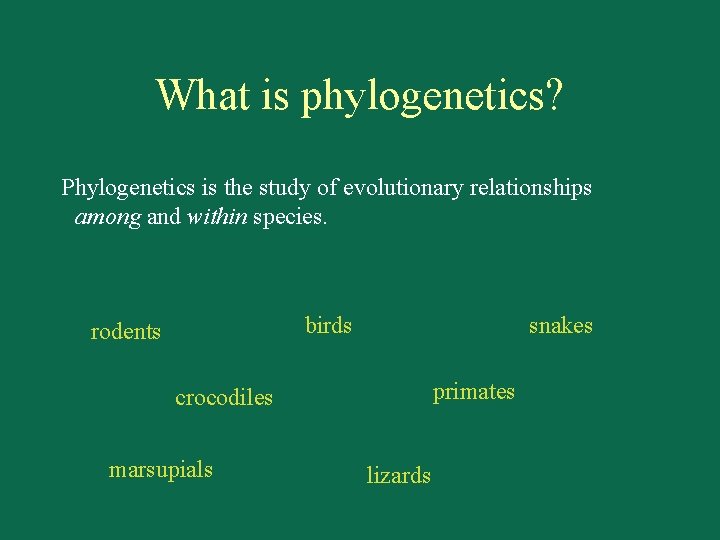
What is phylogenetics? Phylogenetics is the study of evolutionary relationships among and within species. birds rodents snakes primates crocodiles marsupials lizards
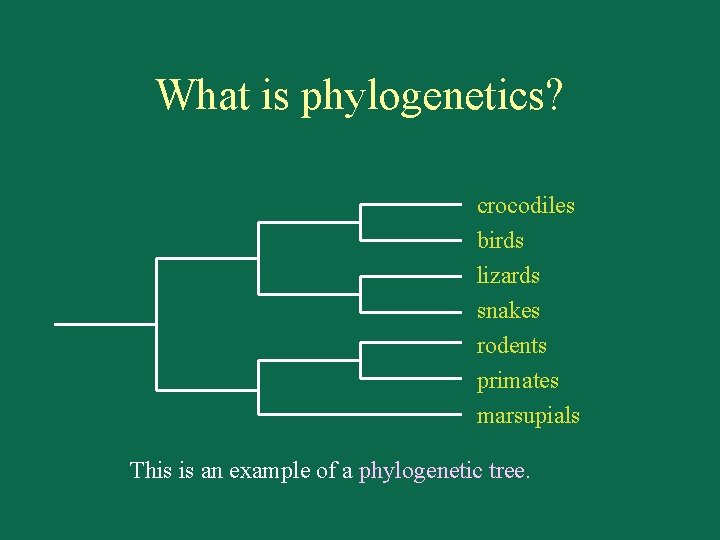
What is phylogenetics? crocodiles birds lizards snakes rodents primates marsupials This is an example of a phylogenetic tree.

Applications of phylogenetics • Forensics: Did a patient’s HIV infection result from an invasive dental procedure performed by an HIV+ dentist? • Conservation: How much gene flow is there among local populations of island foxes off the coast of California? • Medicine: What are the evolutionary relationships among the various prion-related diseases? To be continued…
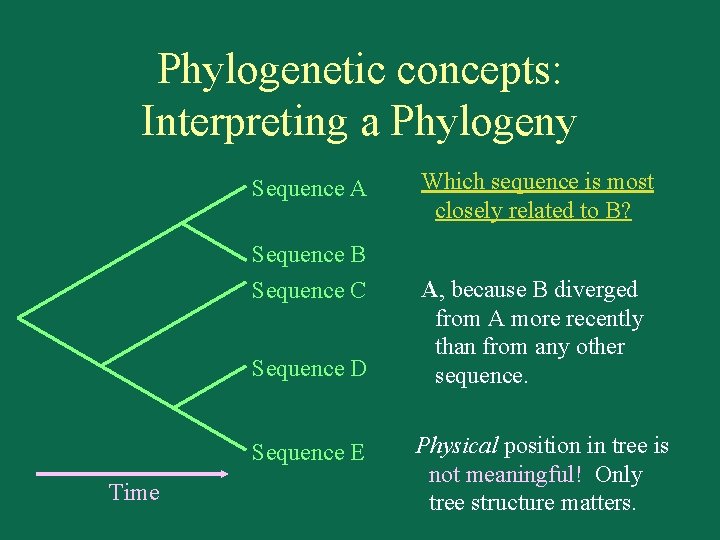
Phylogenetic concepts: Interpreting a Phylogeny Sequence A Sequence B Sequence C Sequence D Sequence E Time Which sequence is most closely related to B? A, because B diverged from A more recently than from any other sequence. Physical position in tree is not meaningful! Only tree structure matters.
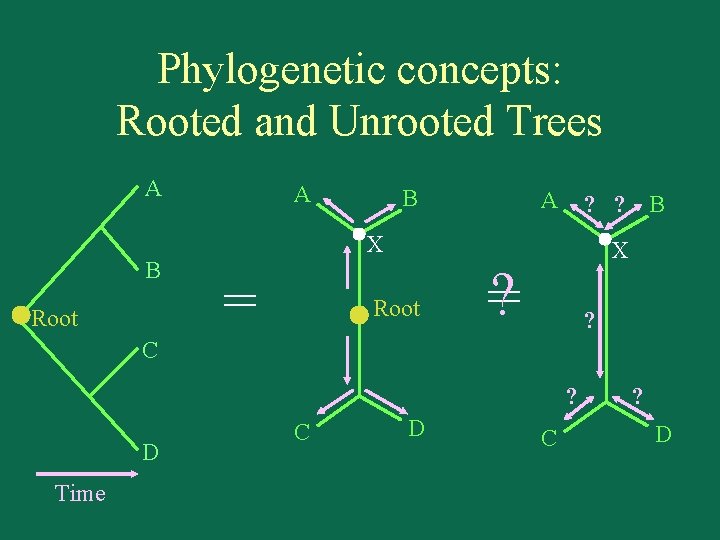
Phylogenetic concepts: Rooted and Unrooted Trees A B Root A B A ? ? B X = Root X ? = ? C ? D Time C D C ? D
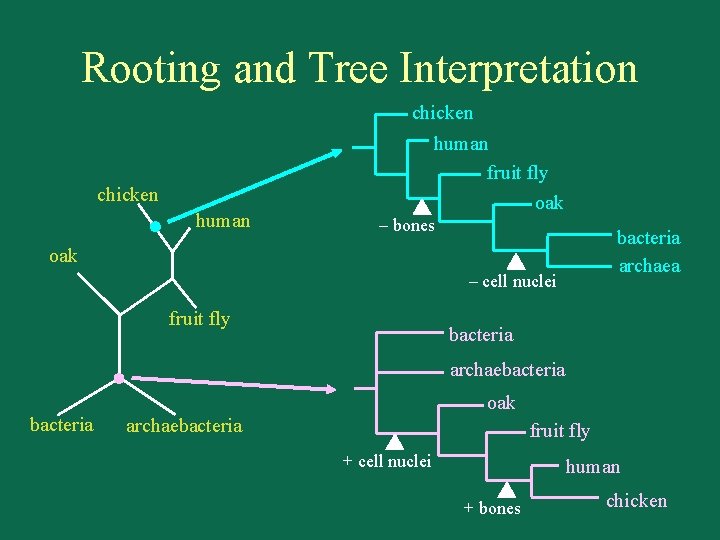
Rooting and Tree Interpretation chicken human fruit fly oak chicken human – bones bacteria archaea oak – cell nuclei fruit fly bacteria archaebacteria oak archaebacteria fruit fly + cell nuclei human + bones chicken
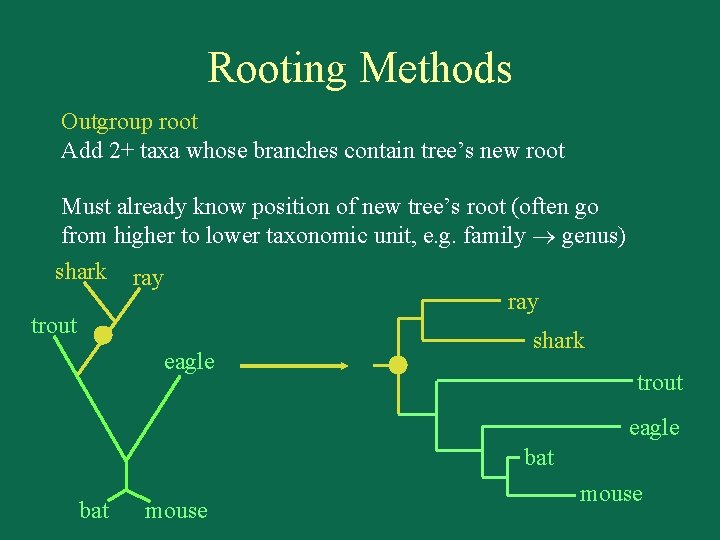
Rooting Methods Outgroup root Add 2+ taxa whose branches contain tree’s new root Must already know position of new tree’s root (often go from higher to lower taxonomic unit, e. g. family genus) shark ray trout shark eagle trout eagle bat mouse
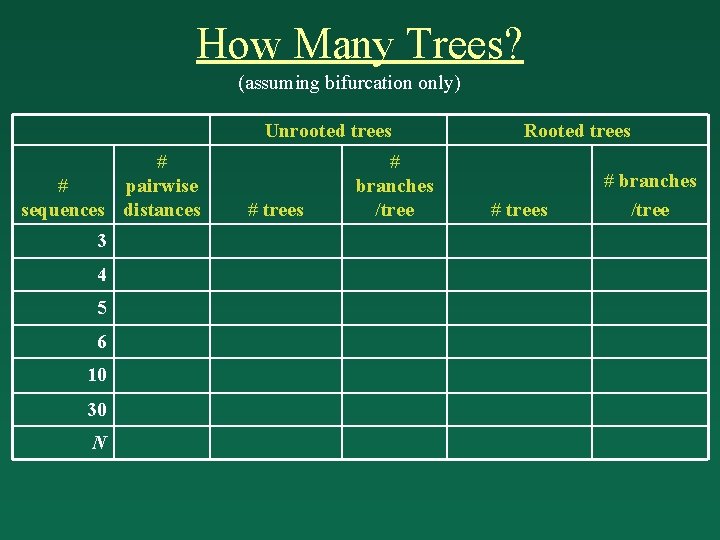
How Many Trees? (assuming bifurcation only) Unrooted trees # # pairwise sequences distances 3 4 5 6 10 30 N # trees # branches /tree Rooted trees # branches /tree
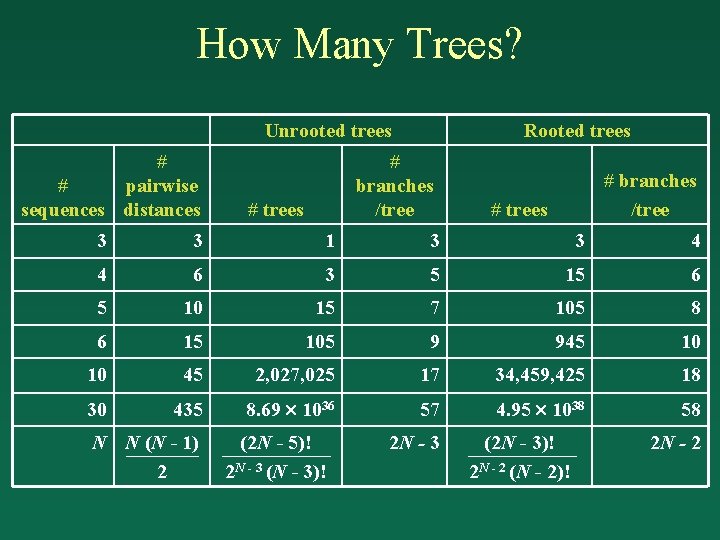
How Many Trees? Unrooted trees # # pairwise sequences distances Rooted trees # branches /tree # trees 3 3 1 3 3 4 4 6 3 5 15 6 5 10 15 7 105 8 6 15 105 9 945 10 10 45 2, 027, 025 17 34, 459, 425 18 30 435 8. 69 1036 57 4. 95 1038 58 N N (N - 1) 2 (2 N - 5)! 2 N - 3 (N - 3)! 2 N - 3 (2 N - 3)! 2 N - 2 (N - 2)! 2 N - 2
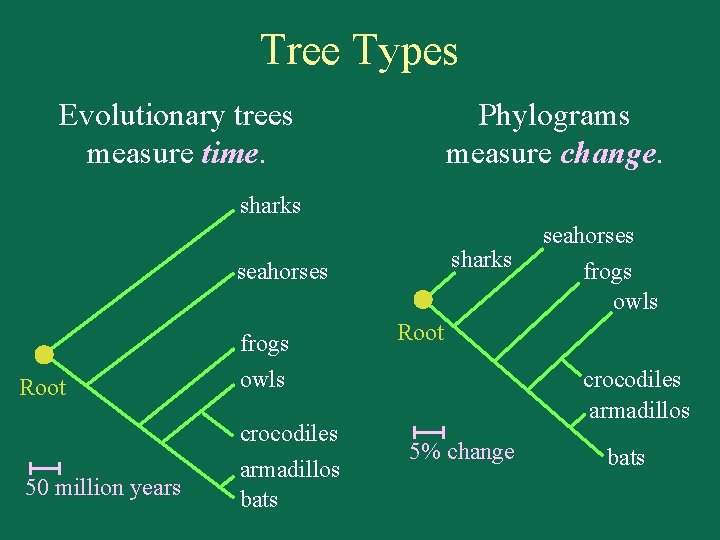
Tree Types Evolutionary trees measure time. Phylograms measure change. sharks seahorses frogs Root 50 million years Root owls crocodiles armadillos bats seahorses frogs owls crocodiles armadillos 5% change bats
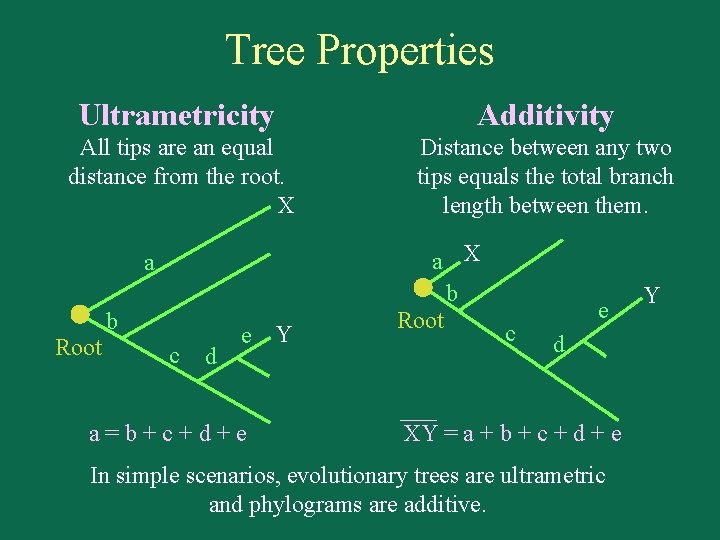
Tree Properties Ultrametricity Additivity All tips are an equal distance from the root. X Distance between any two tips equals the total branch length between them. a Root b c d e Y a=b+c+d+e a X b Root c e d XY = a + b + c + d + e In simple scenarios, evolutionary trees are ultrametric and phylograms are additive. Y
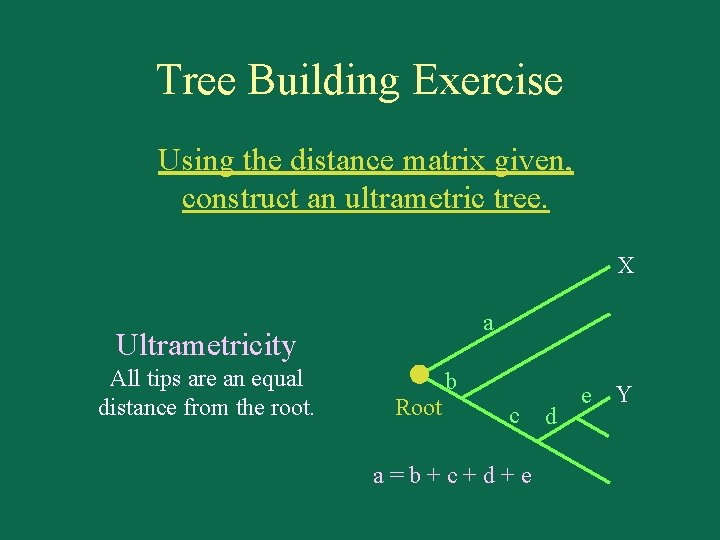
Tree Building Exercise Using the distance matrix given, construct an ultrametric tree. X a Ultrametricity All tips are an equal distance from the root. Root b c a=b+c+d+e d e Y
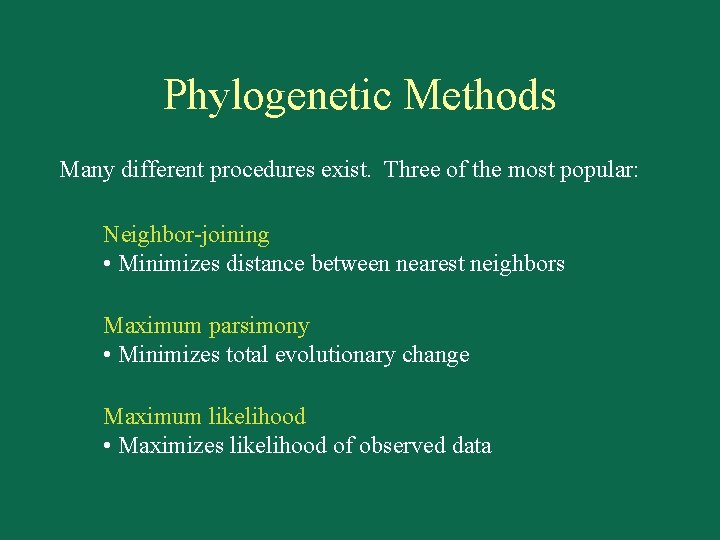
Phylogenetic Methods Many different procedures exist. Three of the most popular: Neighbor-joining • Minimizes distance between nearest neighbors Maximum parsimony • Minimizes total evolutionary change Maximum likelihood • Maximizes likelihood of observed data
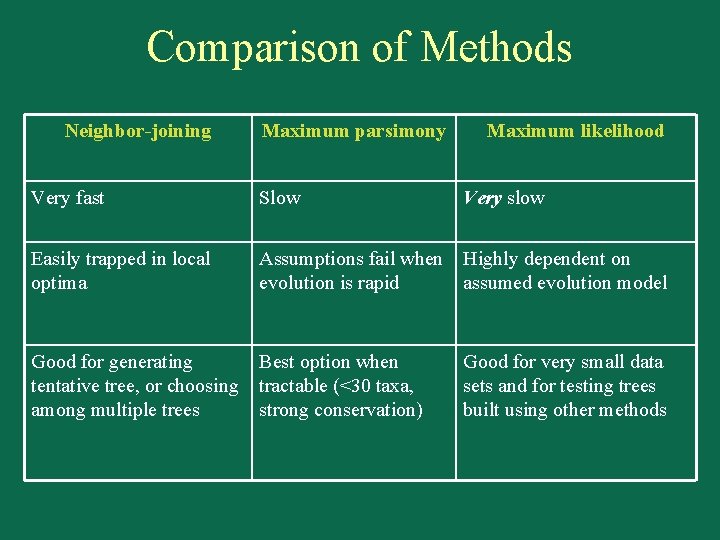
Comparison of Methods Neighbor-joining Maximum parsimony Maximum likelihood Very fast Slow Very slow Easily trapped in local optima Assumptions fail when Highly dependent on evolution is rapid assumed evolution model Good for generating tentative tree, or choosing among multiple trees Best option when tractable (<30 taxa, strong conservation) Good for very small data sets and for testing trees built using other methods
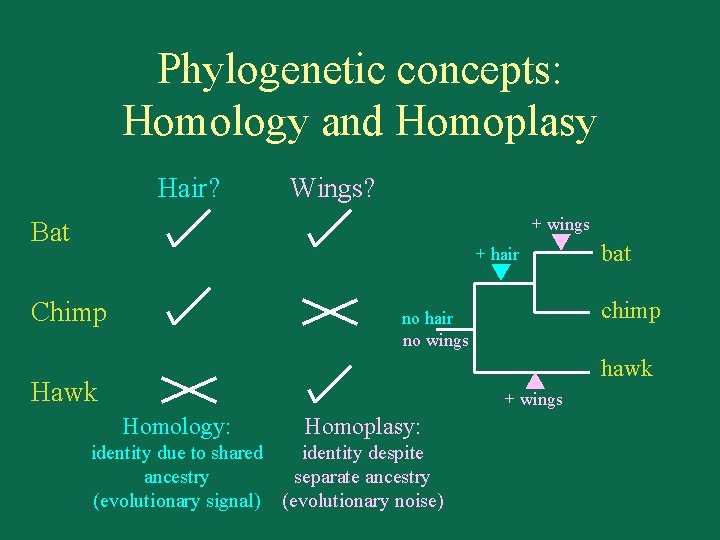
Phylogenetic concepts: Homology and Homoplasy Hair? Wings? + wings Bat + hair Chimp bat chimp no hair no wings hawk Hawk + wings Homology: Homoplasy: identity due to shared identity despite ancestry separate ancestry (evolutionary signal) (evolutionary noise)

Trees are hypotheses about evolutionary history So far, we’ve looked at understanding and formulating these hypotheses. Now, let’s turn our attention to testing them.
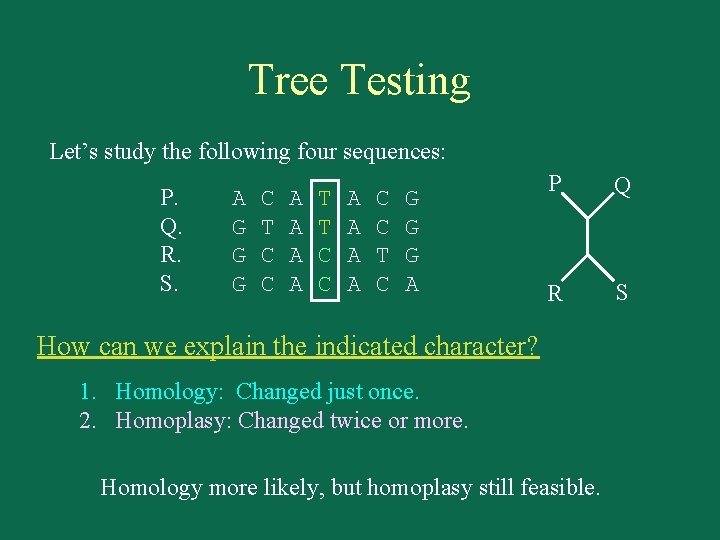
Tree Testing Let’s study the following four sequences: P. Q. R. S. A G G G C T C C A A T T C C A A C C T C G G G A P Q R S How can we explain the indicated character? 1. Homology: Changed just once. 2. Homoplasy: Changed twice or more. Homology more likely, but homoplasy still feasible.
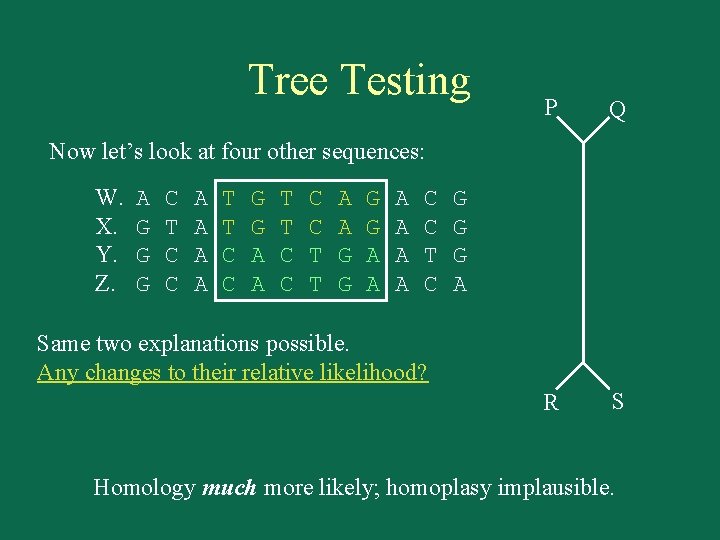
Tree Testing P Q R S Now let’s look at four other sequences: W. X. Y. Z. A G G G C T C C A A T T C C G G A A T T C C T T A A G G A A A C C T C G G G A Same two explanations possible. Any changes to their relative likelihood? Homology much more likely; homoplasy implausible.
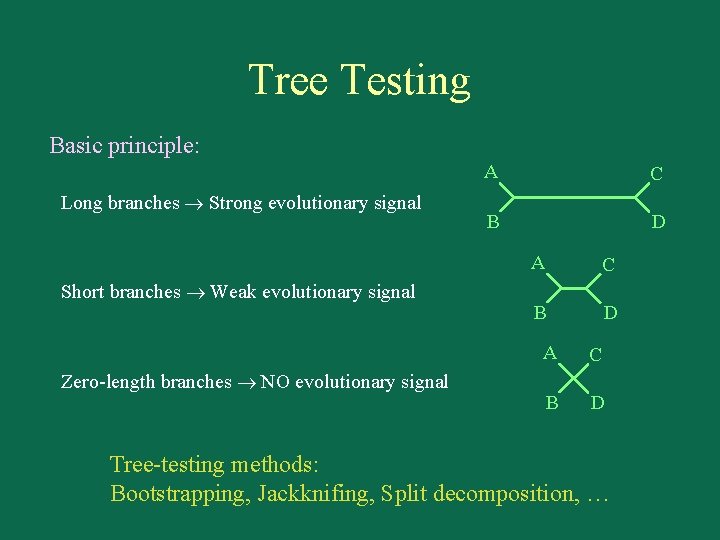
Tree Testing Basic principle: Long branches Strong evolutionary signal Short branches Weak evolutionary signal Zero-length branches NO evolutionary signal A C B D Tree-testing methods: Bootstrapping, Jackknifing, Split decomposition, …

Applications of phylogenetics 1. Forensics Did a patient’s HIV infection result from an invasive dental procedure performed by an HIV+ dentist?
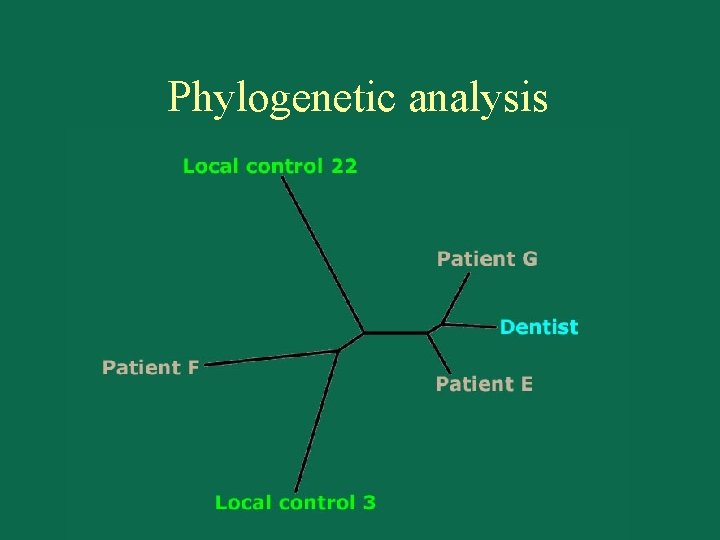
Phylogenetic analysis
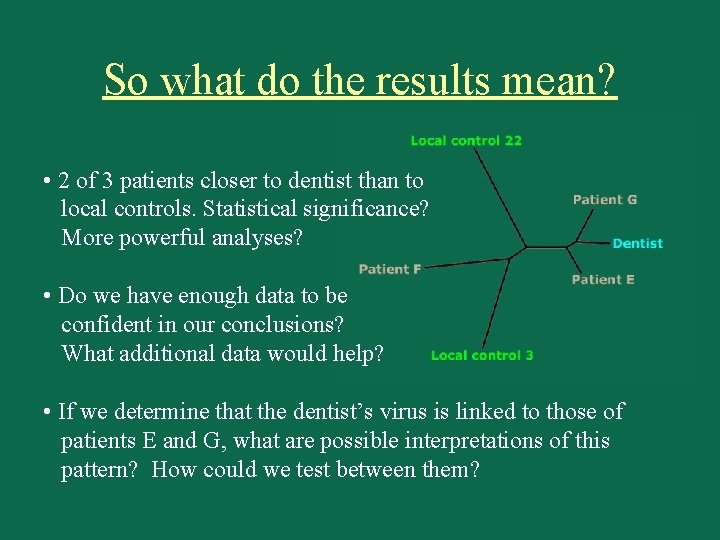
So what do the results mean? • 2 of 3 patients closer to dentist than to local controls. Statistical significance? More powerful analyses? • Do we have enough data to be confident in our conclusions? What additional data would help? • If we determine that the dentist’s virus is linked to those of patients E and G, what are possible interpretations of this pattern? How could we test between them?

Applications of phylogenetics 2. Conservation How much gene flow is there among local populations of island foxes off the coast of California?
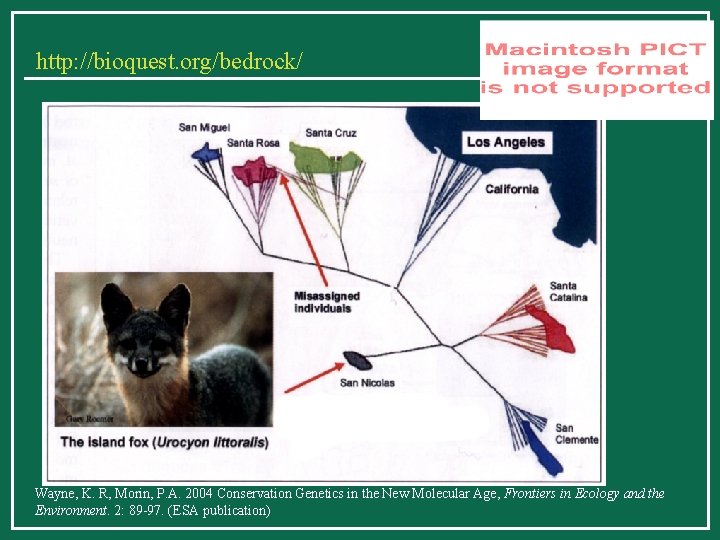
http: //bioquest. org/bedrock/ Wayne, K. R, Morin, P. A. 2004 Conservation Genetics in the New Molecular Age, Frontiers in Ecology and the Environment. 2: 89 -97. (ESA publication)

Applications of phylogenetics 3. Medicine What are the evolutionary relationships among the various prion-related diseases?
Linking Sequence and Structure Enolase
- Slides: 27