The evolution of complexity A primer on evolution



![Ancestral Rhodophyta genes? min. D [min. E] ilv. B (pbs. A) (cpe. B) ilv. Ancestral Rhodophyta genes? min. D [min. E] ilv. B (pbs. A) (cpe. B) ilv.](https://slidetodoc.com/presentation_image/4a843ba7636d03237f56a21a85717787/image-19.jpg)
- Slides: 19

The evolution of complexity


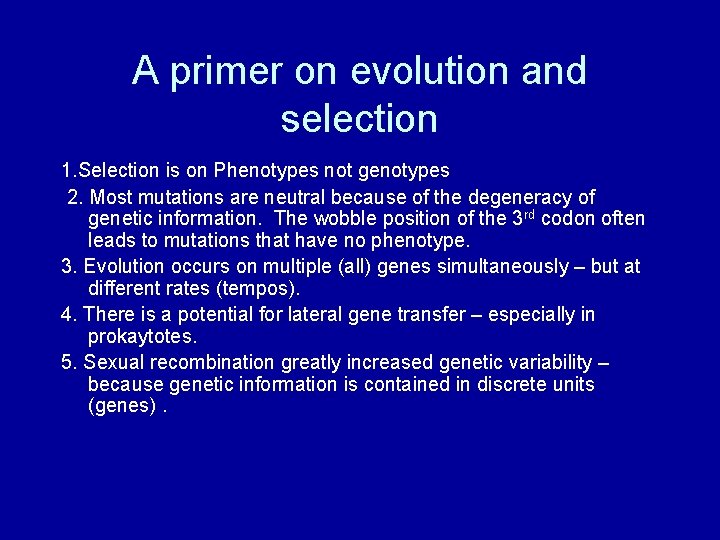
A primer on evolution and selection 1. Selection is on Phenotypes not genotypes 2. Most mutations are neutral because of the degeneracy of genetic information. The wobble position of the 3 rd codon often leads to mutations that have no phenotype. 3. Evolution occurs on multiple (all) genes simultaneously – but at different rates (tempos). 4. There is a potential for lateral gene transfer – especially in prokaytotes. 5. Sexual recombination greatly increased genetic variability – because genetic information is contained in discrete units (genes).


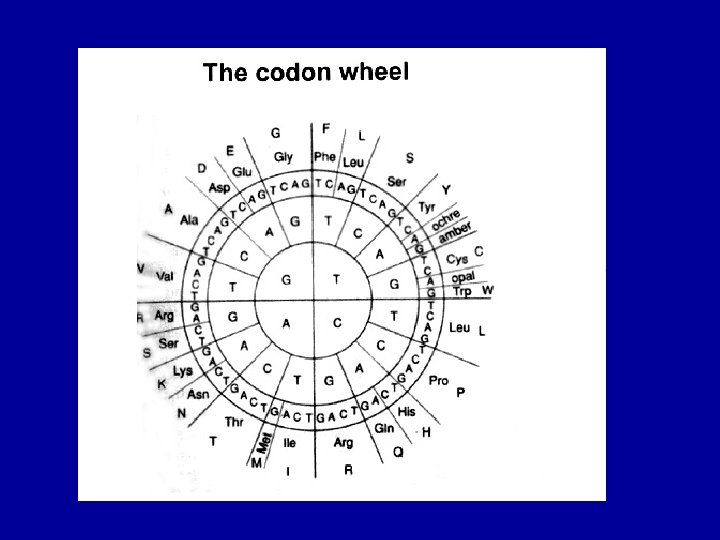
Molecular “clocks” The basic Poisson function for a mutation at any point in a codon d. N/dt (I) = e- kt (kt)I/ I !

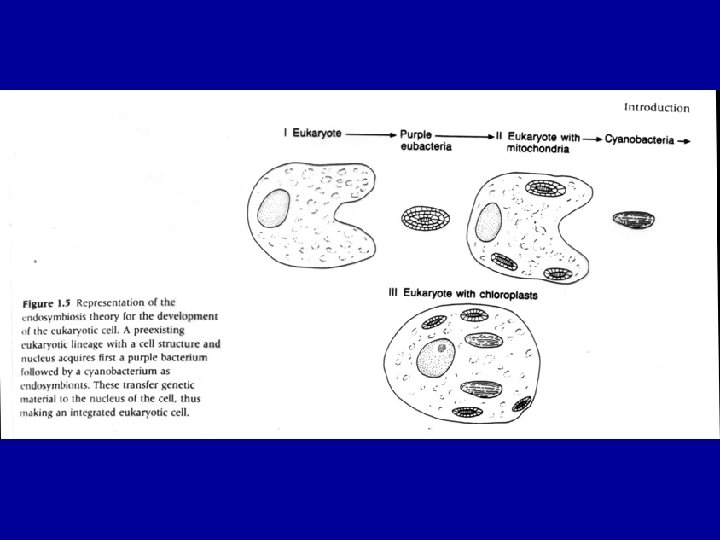
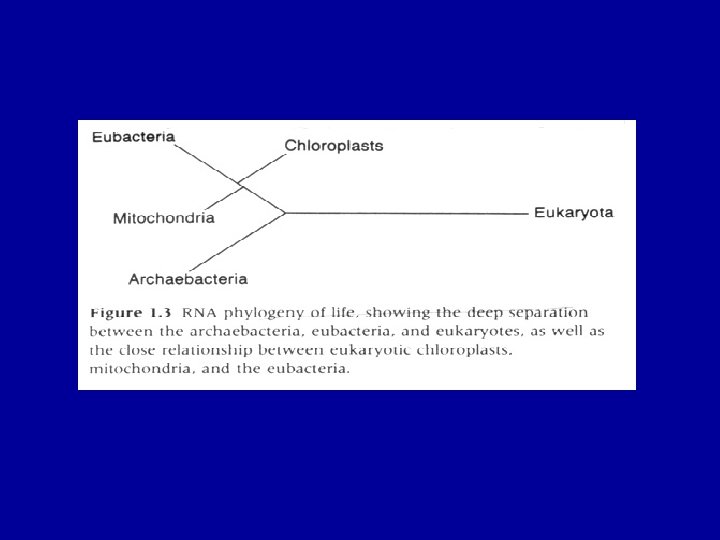
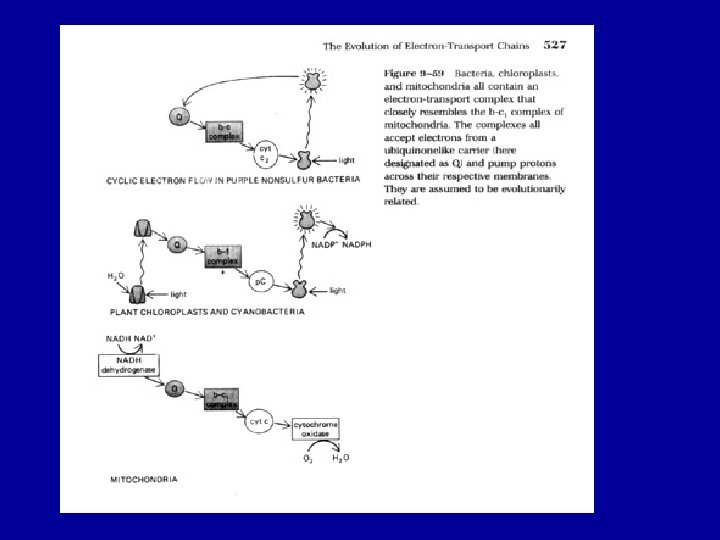
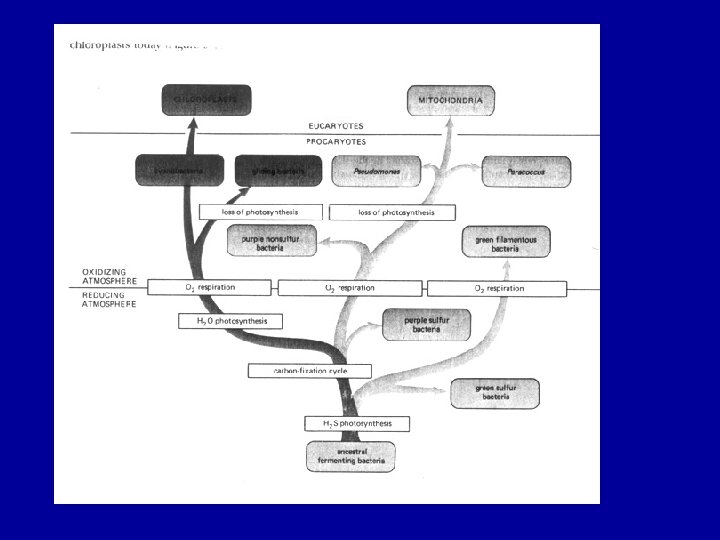
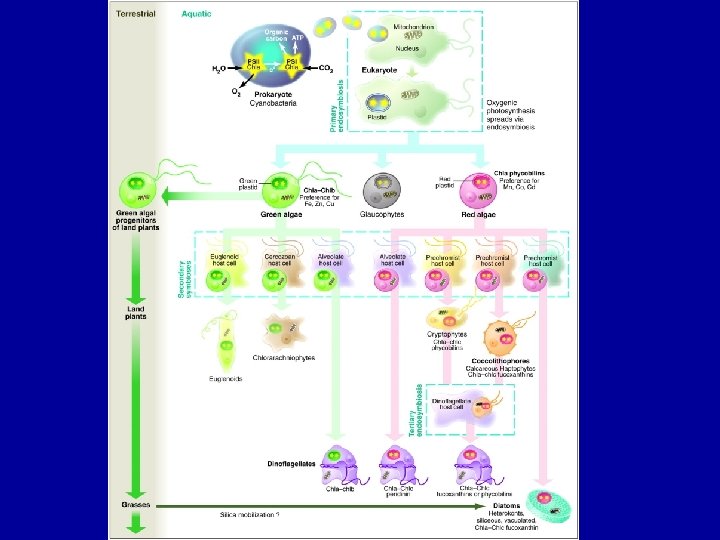
The evolution of Eukaryotes The acquisition of mitochondria and plasids – driving forces and evolutionary consequences Massive lateral gene transfers

Modes of Evolution 1. Horizontal versus lateral gene transfer 2. Selection versus neutral








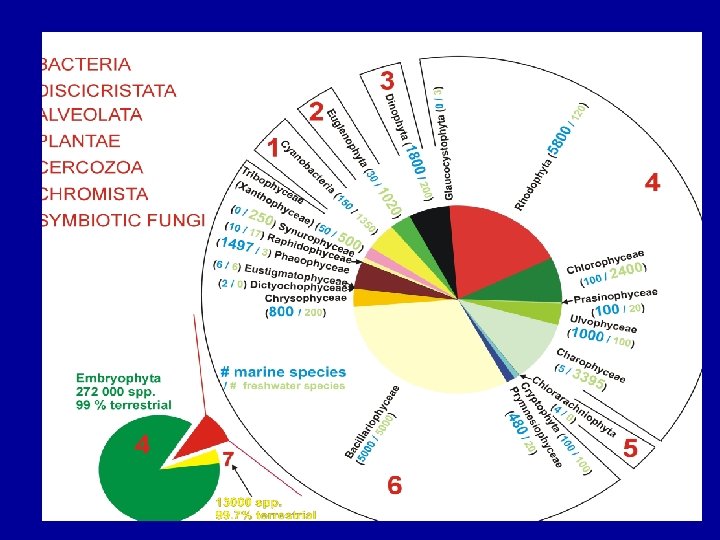
Secondary symbionts • The evolution of modern eukaryotic phytoplankton • Drivers for secondary symbiosis – • retention of fixed N in the host cell (extreme oligotrophy) • plastid could obtain protection from predation (armor)



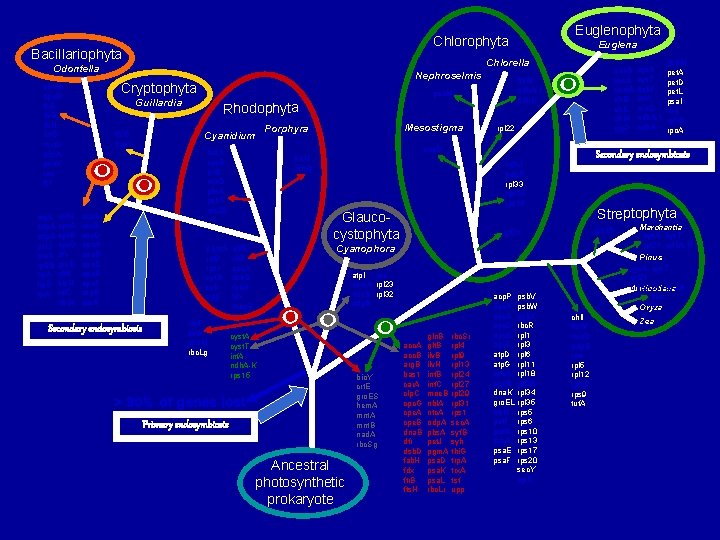
Bacillariophyta ntc. A odp. B pet. J pre. A rpl 28 trp. A trp. G trx. A chl. N cpc. A cpc. B cpc. G dfr gln. B glt. B his. H inf. C nbl. A Euglena Chlorella Odontella Nephroselmis Cryptophyta cem. A cpe. B ftr. B ilv. H inf. B min. D pbs. A psa. K rne tsf Euglenophyta Chlorophyta thi. G bas 1 acc. A acc. B acc. D apc. A apc. B apc. D apc. E apc. F arg. B car. A Rhodophyta Cyanidium clp. P fts. W psb. M rbc. Lg Mesostigma Porphyra rpl 22 acc. D bas 1 cpe. B inf. B min. D pbs. A psb. X rps 20 pgm. A rpl 9 rps 1 syf. B syh upp Secondary endosymbiosis fts. W ndh. A-I ndh. K psa. M Guillardia acc. D ccs. A cem. A chl. B chl. L chl. N clp. P his. H min. D Streptophyta rps 16 rpl 21 Cyanophora chl. B chl. L cpe. A dsb. D fab. H fdx moe. B atp. I rne cem. A rpl 23 min. D rpl 32 odp. B cyst. A cyst. T inf. A ndh. A-K rps 15 > 90% of genes lost Primary endosymbiosis Ancestral photosynthetic prokaryote bio. Y crt. E gro. ES hem. A mnt. B nad. A rbc. Sg acc. A acc. B arg. B bas 1 car. A clp. C cpc. G cpe. A cpe. B dna. B dfr dsb. D fab. H fdx ftr. B fts. H gln. B glt. B ilv. H inf. B inf. C moe. B nbl. A ntc. A odp. A pbs. A pet. J pgm. A psa. D psa. K psa. L rbc. Lr rbc. Sr rpl 4 rpl 9 rpl 13 rpl 24 rpl 27 rpl 29 rpl 31 rps 1 sec. A syf. B syh thi. G trp. A trx. A tsf upp acp. P apc. A apc. B apc. D apc. E apc. F atp. D atp. G cpc. A cpc. B dna. K gro. EL his. H pet. F pet. M pre. A psa. E psa. F psb. V psb. W psb. X rbc. R rpl 1 rpl 3 rpl 6 rpl 11 rpl 18 rpl 28 rpl 34 rpl 35 rps 6 rps 10 rps 13 rps 17 rps 20 sec. Y trp. G psb. M pet. A pet. D pet. L psa. I rne rpl 19 rpo. A Secondary endosymbiosis ndh. J odp. B rpl 33 rps 15 rps 16 Glaucocystophyta cys. A cys. T fts. W inf. A min. D ndh. A-I ndh. K Marchantia cys. A, cys. T rpl 21, ndh. A-K Pinus cys. A, cys. T, rpl 21 chl. B, L, N, psa. M Nicotiana acc D Oryza chl. I fts. W min. D odp. B rne rpl 5 rpl 12 rpl 19 rps 9 tuf. A Zea
![Ancestral Rhodophyta genes min D min E ilv B pbs A cpe B ilv Ancestral Rhodophyta genes? min. D [min. E] ilv. B (pbs. A) (cpe. B) ilv.](https://slidetodoc.com/presentation_image/4a843ba7636d03237f56a21a85717787/image-19.jpg)
Ancestral Rhodophyta genes? min. D [min. E] ilv. B (pbs. A) (cpe. B) ilv. H psa. K ftr. B (inf. B) tsf Cryptophyta Guillardia [acc. D] [odp. B] cem. A [(rne)] Euglenophyta Euglena atp. I rpl 23 rpl 32 Bacillariophyta Odontella thi. G (bas 1) acc. A acc. B arg. B car. A cpc. G (cpe. A) dfr (dsb. D) (fab. H) (fdx) gln. B glt. B inf. C moe. B nbl. A ntc. A odp. A pet. J (pgm. A) rpl 9 (rps 1) Rhodophyta (syf. B) (syh) trp. A trx. A (upp) cbb. X clp. C dna. B fts. H psa. D psa. L rbc. Sr rpl 4 rpl 13 rpl 24 rpl 27 rpl 29 rpl 31 sec. A acp. A/P atp. D atp. G dna. K gro. EL pet. F pet. M psa. E psa. F psb. V psb. W (psb. X) rbc. R apc. A apc. B apc. D apc. E rpl 1 rpl 3 rpl 6 rpl 11 rpl 18 rpl 21 rpl 34 rpl 35 rps 6 rps 10 rps 13 rps 17 (rps 20) sec. Y atp. A atp. B atp. E atp. F atp. H chl. I pet. B pet. G psa. A psa. B psa. C psa. J [psa. M] psb. A psb. B ccs. A pet. D pet. L psa. I rpl 19 [rpl 33] [rps 16] rpo. A psb. C psb. D psb. E psb. F psb. H psb. I psb. J psb. K psb. L psb. N psb. T rbc. Lg/r rpl 2 rpl 5 rpl 12 rpl 14 rpl 16 rpl 20 [rpl 22] rpl 36 rps 2 rps 3 rps 4 rps 7 rps 8 rps 9 rps 11 rps 12 rps 14 rps 18 rps 19 rpo. B rpo. C 1 rpo. C 2 tuf. A [rdp. O] cys. A cys. T inf. A [I-Cvu. I] [ndh. A] [ndh. B] [ndh. C] [ndh. D] [ndh. E] [ndh. F] [ndh. G] [ndh. H] [ndh. I] [ndh. J] [ndh. K] [rps 15] chl. B (chl. L) chl. N apc. F cpc. A pre. A cpc. B rpl 28 (his. H) trp. G Glaucocystophyceae Cyanophora bio. Y crte gro. ES hem. A clp. P [fts. W] psb. M mnt. A mnt. B nad. A rbc. Sg Chlorophyta