The Eukaryotic Cell Cycle molecules mechanisms modules and





- Slides: 26


The Eukaryotic Cell Cycle: molecules, mechanisms, modules and mathematical models Béla Novák Molecular Network Dynamics Research Group Hungarian Academy of Sciences and Budapest University of Technology and Economics Collaborator: John J. Tyson Department of Biology Virginia Polytechnic Institute and State University

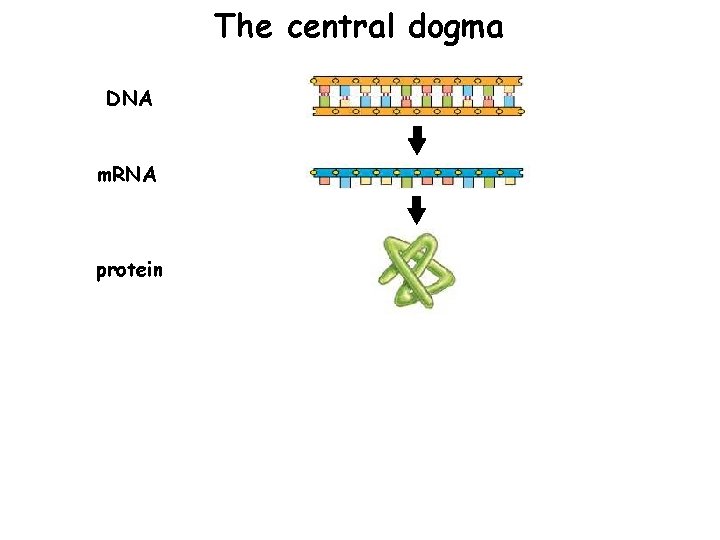
The central dogma DNA m. RNA protein

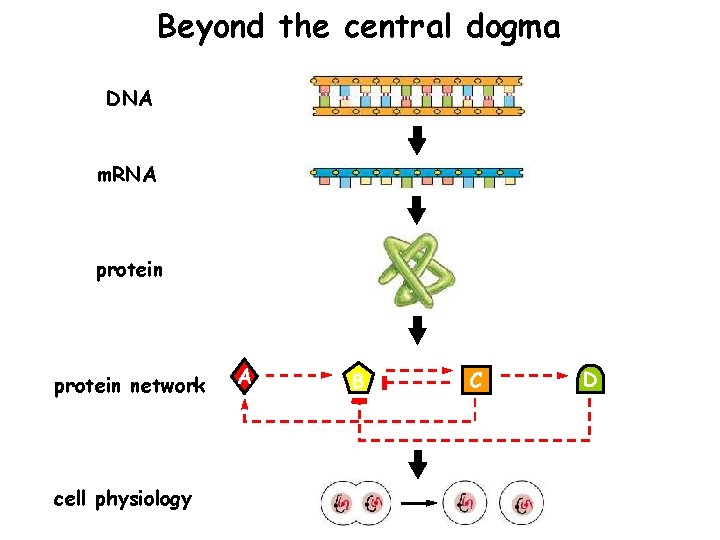
Beyond the central dogma DNA m. RNA protein network cell physiology A B C D

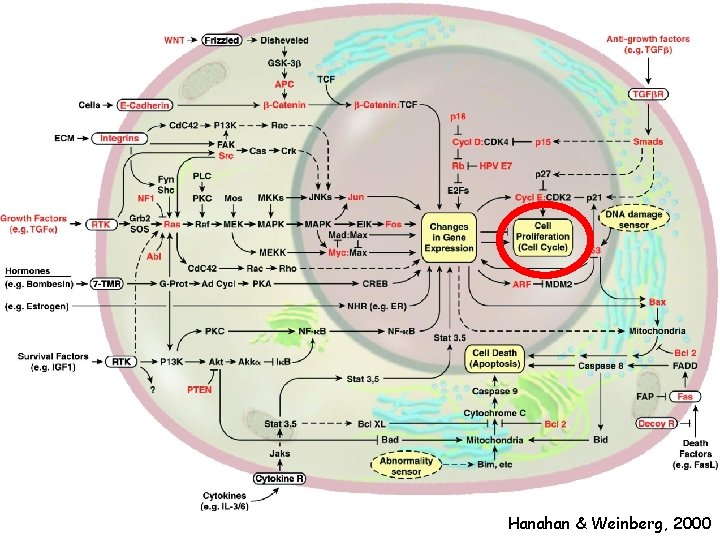
Hanahan & Weinberg, 2000

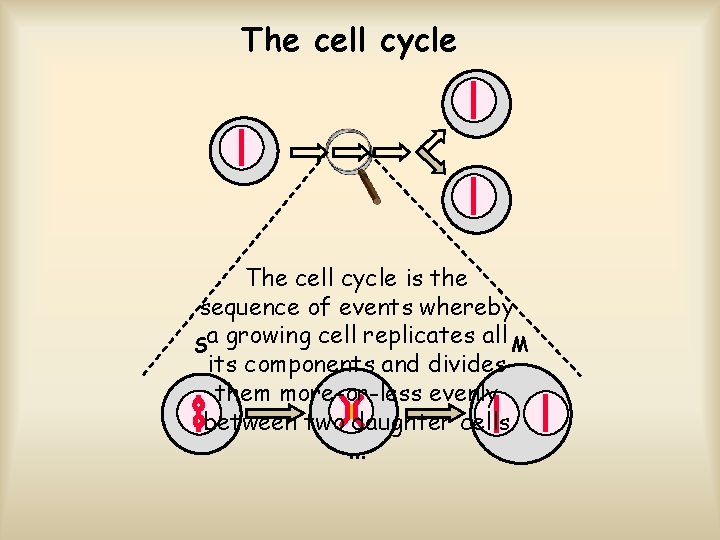
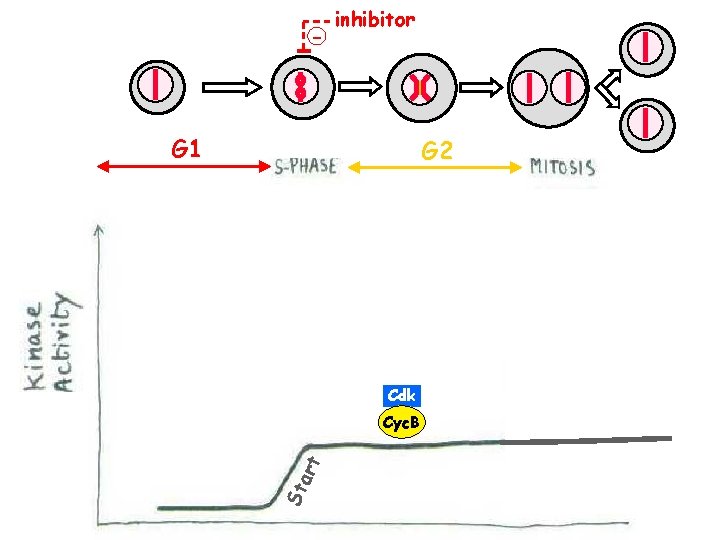
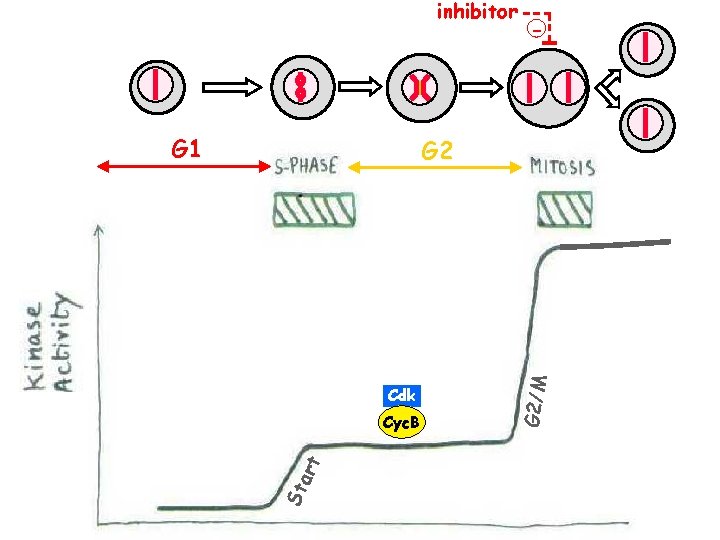
The cell cycle is the sequence of events whereby Sa growing cell replicates all M its components and divides them more-or-less evenly between two daughter cells. . .

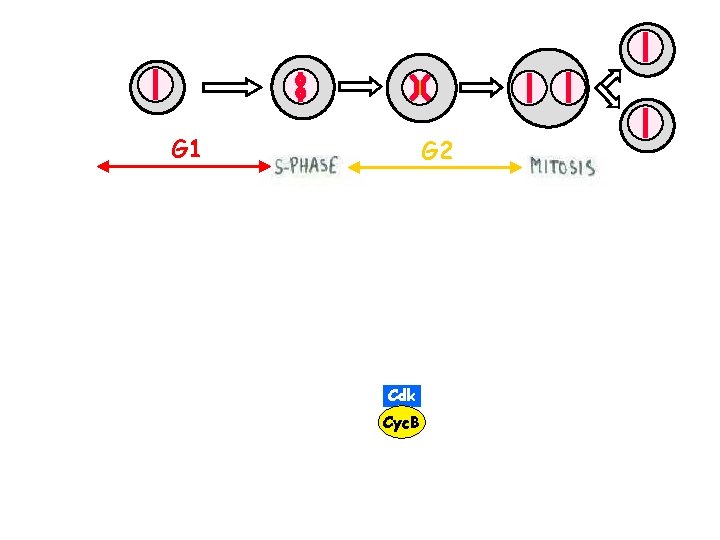
G 1 G 2 Cdk Cyc. B

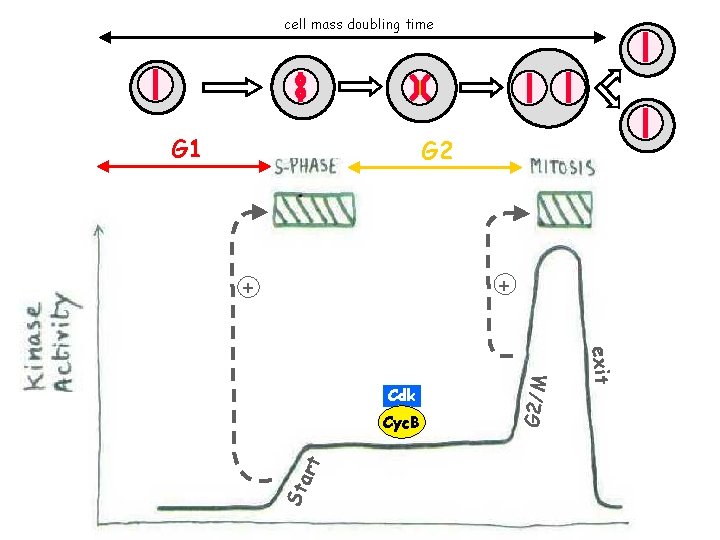
cell mass doubling time G 1 G 2 St art Cyc. B exit Cdk G 2/M + +

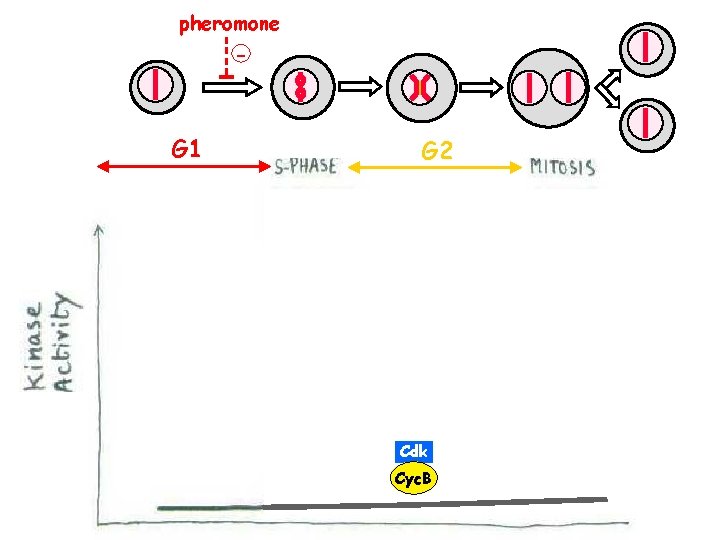
pheromone - G 1 G 2 + Cdk Cyc. B

- inhibitor G 1 G 2 Cdk St art Cyc. B

inhibitor G 1 - Cdk St art Cyc. B G 2/M G 2

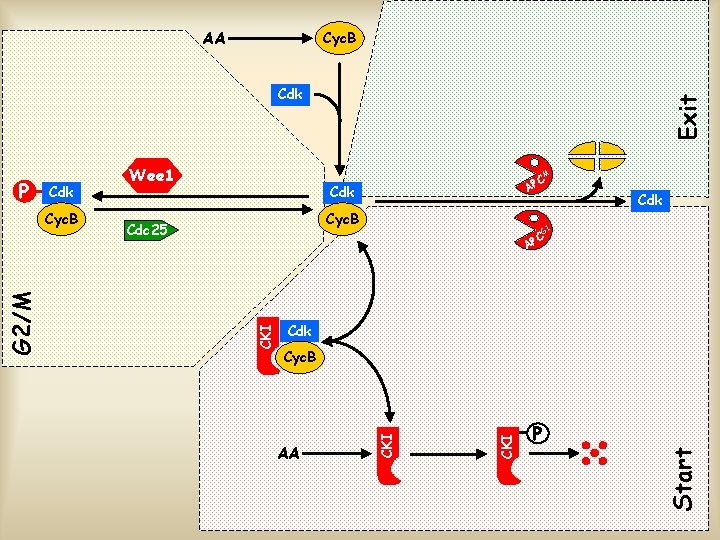
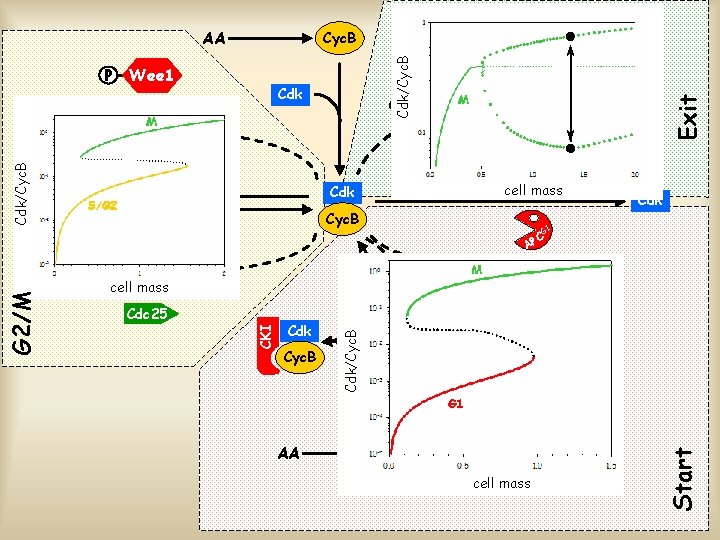
AA Cyc. B Cdk Cyc. B Cdc 25 Cdk G 1 C AP Cdk Cyc. B P Start AA CKI G 2/M Cyc. B C AP CKI Cdk M CKI P Wee 1 Exit Cdk

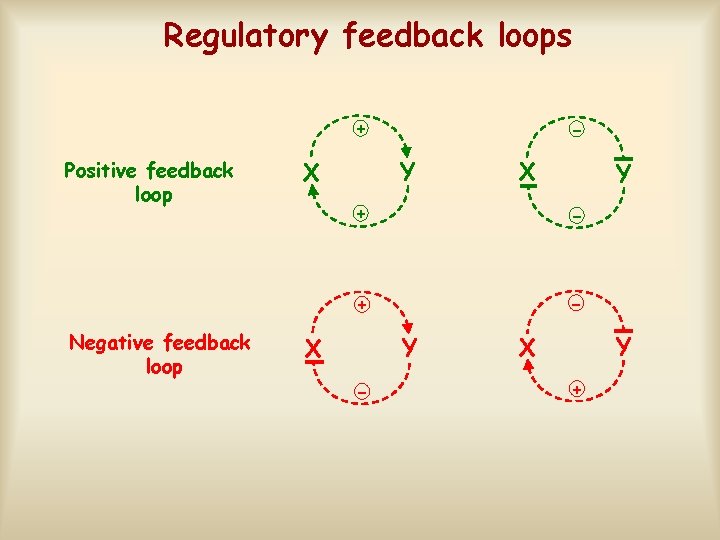
Regulatory feedback loops + Positive feedback loop Negative feedback loop - Y X X Y + - Y X +

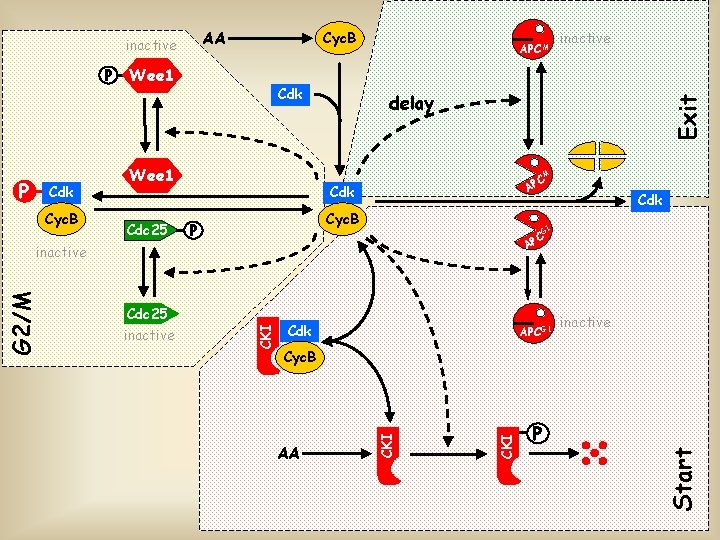
AA P Cdk Cyc. B Wee 1 Cdk Wee 1 Cdc 25 APCM delay M C AP Cdk Cyc. B P G 1 Cdk APCG 1 inactive AA CKI Cyc. B CKI Cdc 25 CKI G 2/M Cdk C AP inactive P Start P Cyc. B Exit inactive

? fission yeast budding yeast

Mutants in Fission Yeast gene viability trait cdc 2 cdc 13 rum 1 ste 9 wee 1 cdc 25 - no no yes yes no block in G 1 and G 2 endoreplication sterile small cells block in G 2 cdc 2 op cdc 13 op rum 1 op ste 9 op wee 1 op cdc 25 op yes no no yes wt wt endoreplication large cells small cells wee 1 - rum 1 wee 1 - cdc 25 op no yes no extremely small quantized cycles mitotic catastrophe

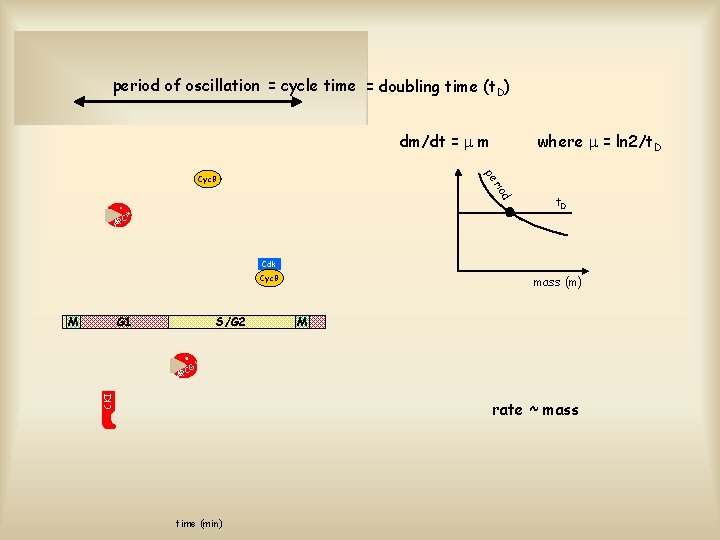
period of oscillation = cycle time = doubling time (t. D) dm/dt = m where = ln 2/t. D pe d rio Cyc. B t. D M C AP Cdk Cyc. B M G 1 S/G 2 mass (m) M 1 CG CKI AP rate ~ mass time (min)

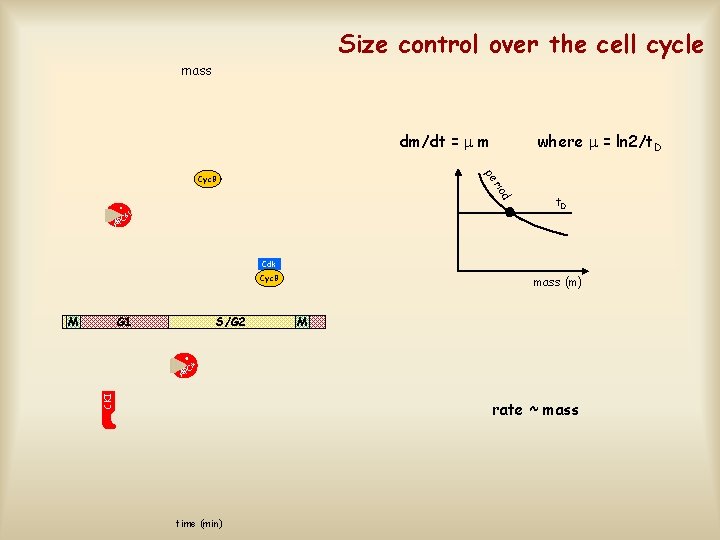
Size control over the cell cycle mass dm/dt = m where = ln 2/t. D pe d rio Cyc. B G 1 t. D C AP Cdk Cyc. B M G 1 S/G 2 mass (m) M M C CKI AP rate ~ mass time (min)

Inside view ?

AA Cdc 25 Cdk/Cyc. B Exit M C AP cell mass Cdk Cyc. B P Cdk G 1 C AP cell mass Cdk Cyc. B AA APCG 1 cell mass P Start Cdc 25 CKI Cyc. B delay CKI Cdk Wee 1 APCM Cdk/Cyc. B G 2/M P Wee 1 CKI Cdk/Cyc. B P Cyc. B

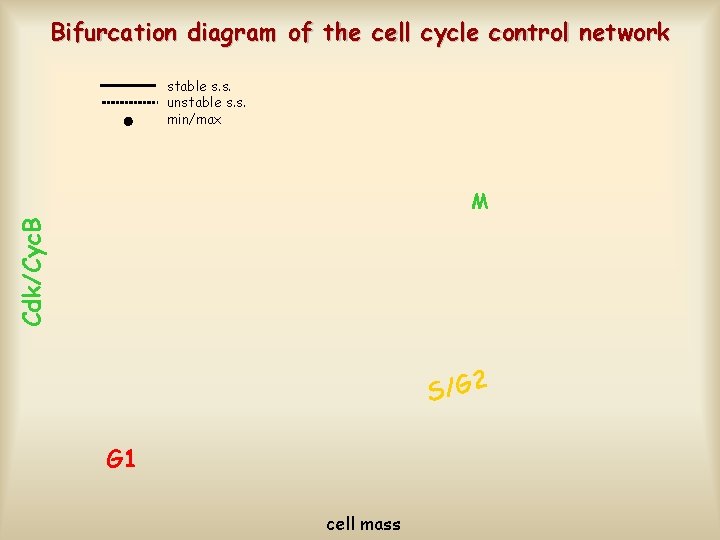
Bifurcation diagram of the cell cycle control network stable s. s. unstable s. s. min/max Cdk/Cyc. B M S/G 2 G 1 cell mass

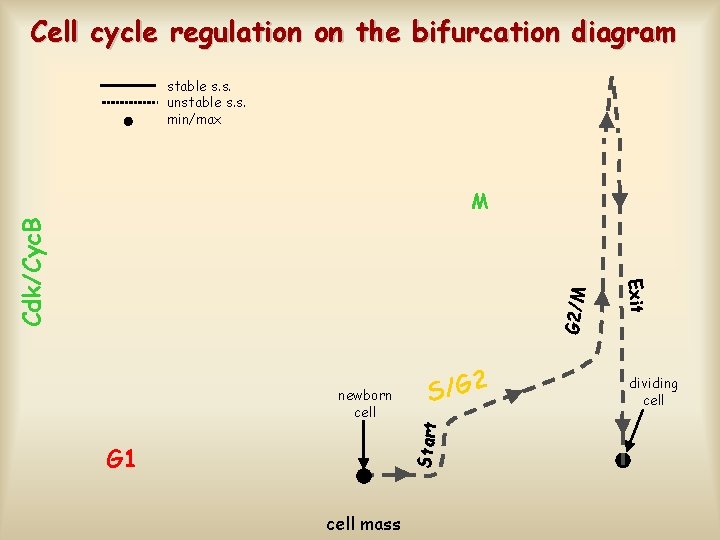
Cell cycle regulation on the bifurcation diagram stable s. s. unstable s. s. min/max G 2/M S/G 2 Start newborn cell G 1 cell mass Exit Cdk/Cyc. B M dividing cell

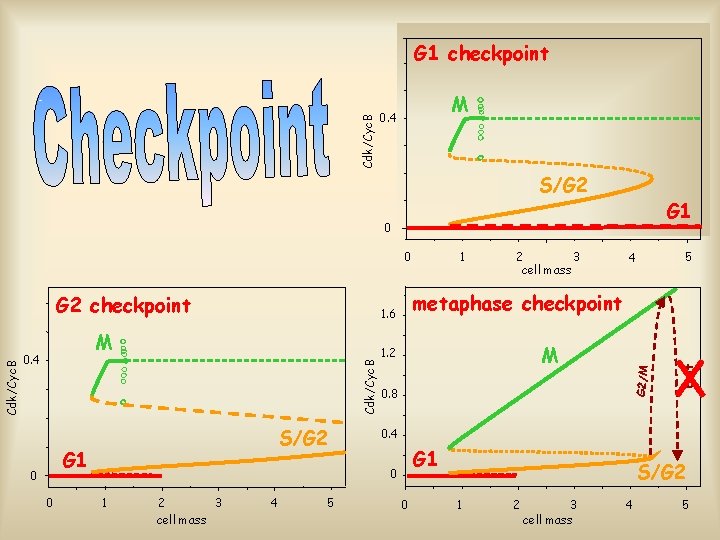
M 0. 4 S/G 2 G 1 0 0 M 1. 2 0. 8 0. 4 G 1 0 1 2 3 cell mass 4 5 5 4 G 2/M M 0. 4 2 3 cell mass metaphase checkpoint 1. 6 Cdk/Cyc. B G 2 checkpoint 1 Exit Cdk/Cyc. B G 1 checkpoint 0 S/G 2 1 2 3 cell mass 4 5

Jason Zwolak Attila Csikász-Nagy Béla Győrffy Ákos John Attila Tyson Sveiczer Tóth Laurence Calzone Kathy Chen Chris Hong Andrea Ciliberto

Homepage: http: //www. cellcycle. bme. hu/ Supporting agencies: Hungarian Academy of Sciences

Modelling the cell division cycle Béla Novák Molecular Network Dynamics Research Group Hungarian Academy of Sciences and Budapest University of Technology and Economics Frank Uhlmann Paul Nurse