The Equipment LCMSMS Fluorescence detector Micromass Quattro Ultima


- Slides: 17

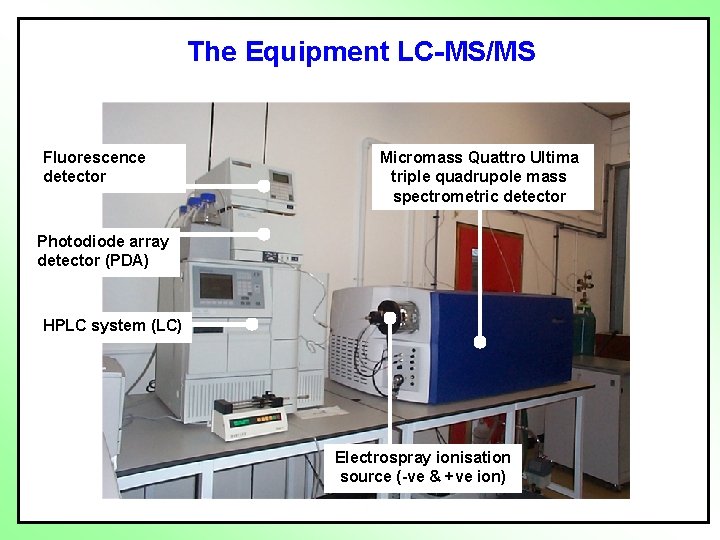
The Equipment LC-MS/MS Fluorescence detector Micromass Quattro Ultima triple quadrupole mass spectrometric detector Photodiode array detector (PDA) HPLC system (LC) Electrospray ionisation source (-ve & +ve ion)

A little bit about triple quadrupole mass spectrometry.

A little bit about electrospray ionisation.

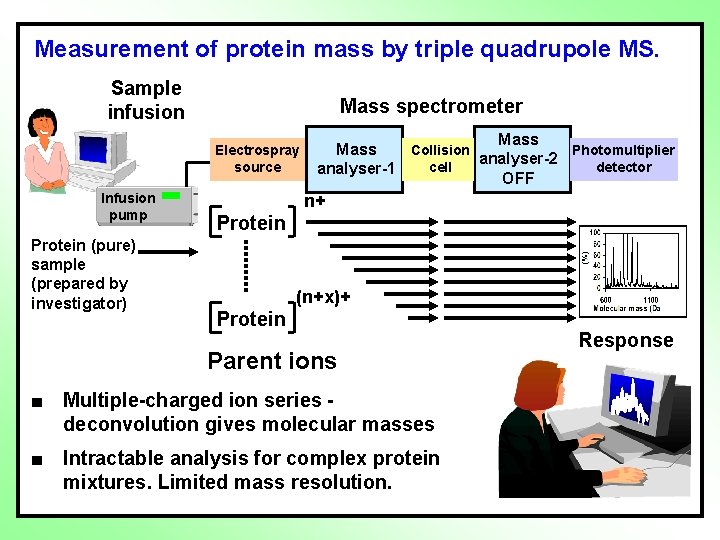
Measurement of protein mass by triple quadrupole MS. Sample infusion Mass spectrometer Electrospray source Infusion pump Protein (pure) sample (prepared by investigator) Mass Collision Photomultiplier analyser-2 cell detector analyser-1 OFF n+ Protein (n+x)+ Protein Parent ions ■ Multiple-charged ion series deconvolution gives molecular masses ■ Intractable analysis for complex protein mixtures. Limited mass resolution. Response

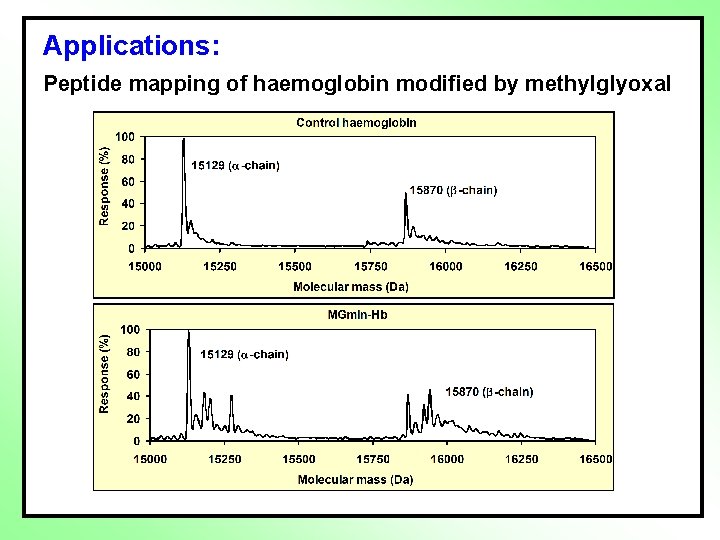
Applications: Peptide mapping of haemoglobin modified by methylglyoxal

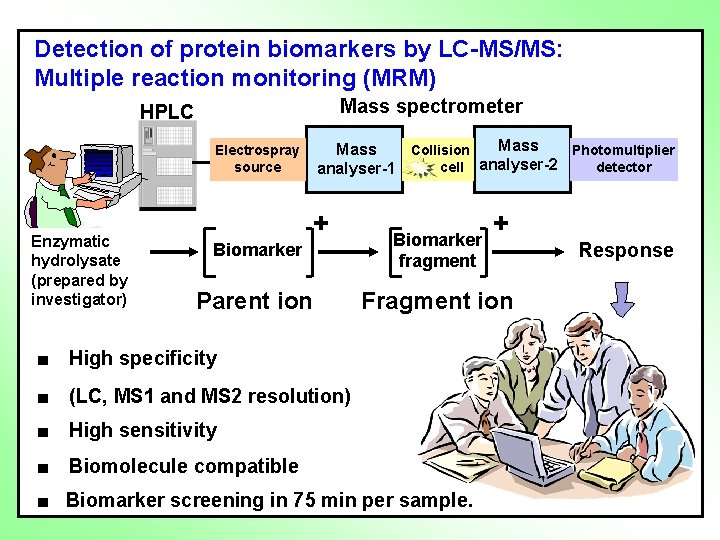
Detection of protein biomarkers by LC-MS/MS: Multiple reaction monitoring (MRM) Mass spectrometer HPLC Electrospray source Enzymatic hydrolysate (prepared by investigator) Mass Collision Photomultiplier cell analyser-2 detector analyser-1 + + Biomarker fragment Parent ion Fragment ion ■ High specificity ■ (LC, MS 1 and MS 2 resolution) ■ High sensitivity ■ Biomolecule compatible ■ Biomarker screening in 75 min per sample. Response

Advanced glycation endproducts LC-MS/MS with stable isotope-substituted internal standards

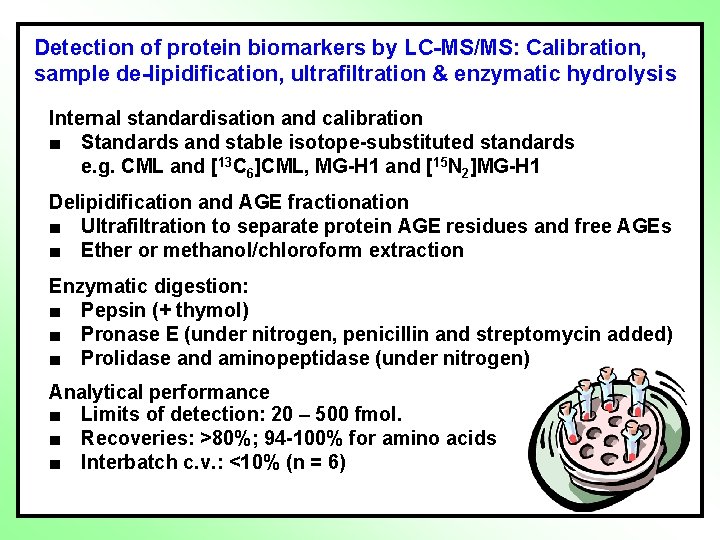
Detection of protein biomarkers by LC-MS/MS: Calibration, sample de-lipidification, ultrafiltration & enzymatic hydrolysis Internal standardisation and calibration ■ Standards and stable isotope-substituted standards e. g. CML and [13 C 6]CML, MG-H 1 and [15 N 2]MG-H 1 Delipidification and AGE fractionation ■ Ultrafiltration to separate protein AGE residues and free AGEs ■ Ether or methanol/chloroform extraction Enzymatic digestion: ■ Pepsin (+ thymol) ■ Pronase E (under nitrogen, penicillin and streptomycin added) ■ Prolidase and aminopeptidase (under nitrogen) Analytical performance ■ Limits of detection: 20 – 500 fmol. ■ Recoveries: >80%; 94 -100% for amino acids ■ Interbatch c. v. : <10% (n = 6)

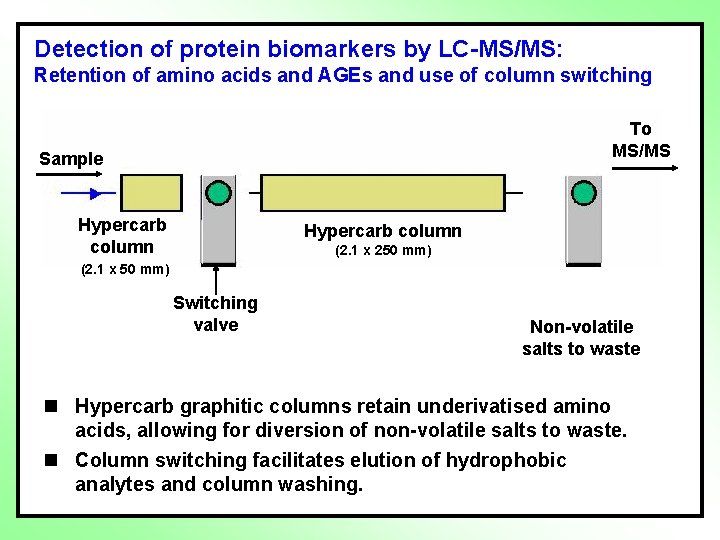
Detection of protein biomarkers by LC-MS/MS: Retention of amino acids and AGEs and use of column switching To MS/MS Sample Hypercarb column (2. 1 x 250 mm) (2. 1 x 50 mm) Switching valve Non-volatile salts to waste n Hypercarb graphitic columns retain underivatised amino acids, allowing for diversion of non-volatile salts to waste. n Column switching facilitates elution of hydrophobic analytes and column washing.

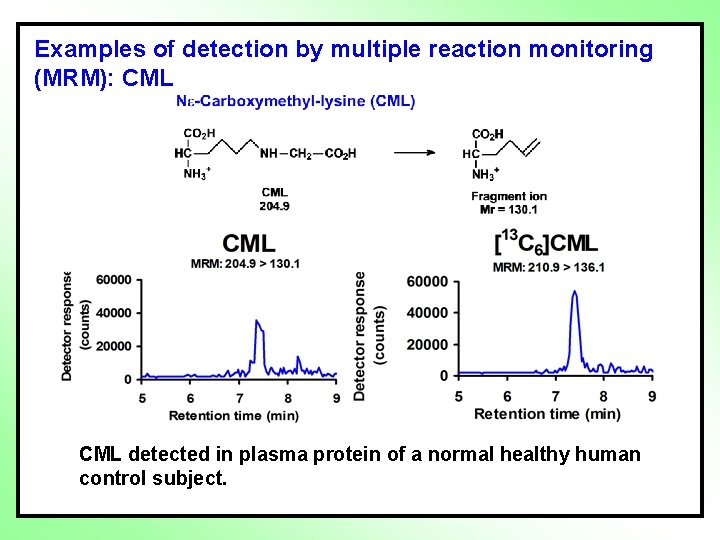
Examples of detection by multiple reaction monitoring (MRM): CML detected in plasma protein of a normal healthy human control subject.

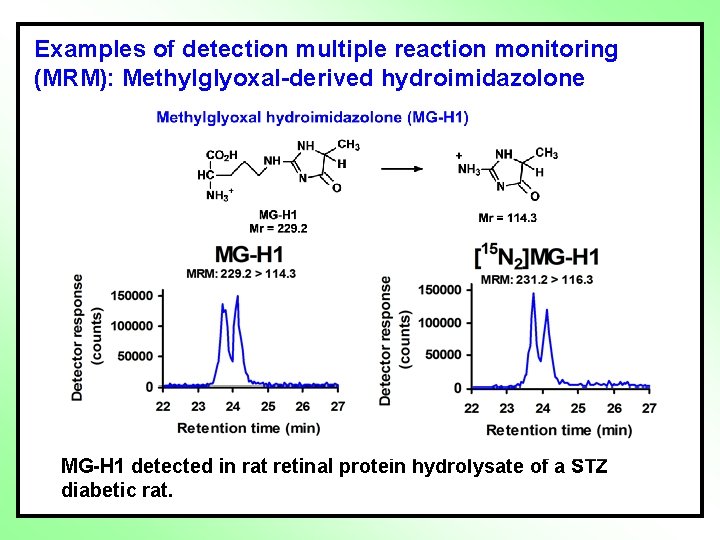
Examples of detection multiple reaction monitoring (MRM): Methylglyoxal-derived hydroimidazolone MG-H 1 detected in rat retinal protein hydrolysate of a STZ diabetic rat.

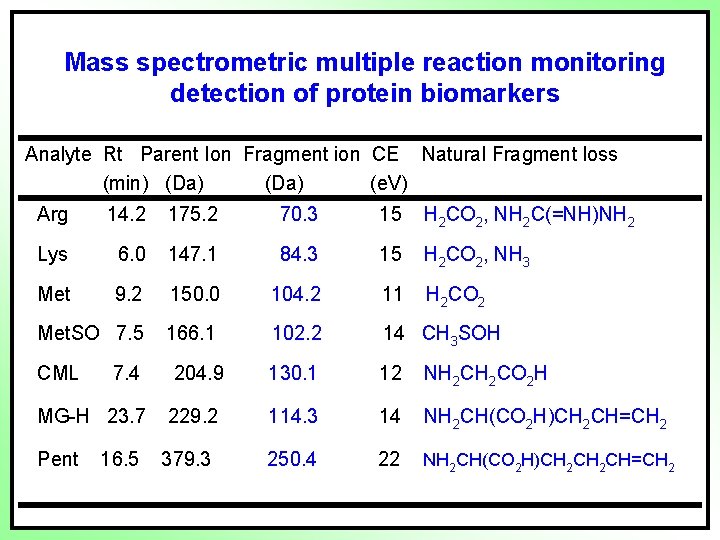
Mass spectrometric multiple reaction monitoring detection of protein biomarkers Analyte Rt Parent Ion Fragment ion CE Natural Fragment loss (min) (Da) (e. V) Arg 14. 2 175. 2 70. 3 15 H 2 CO 2, NH 2 C(=NH)NH 2 Lys 6. 0 147. 1 84. 3 15 H 2 CO 2, NH 3 Met 9. 2 150. 0 104. 2 11 H 2 CO 2 Met. SO 7. 5 166. 1 102. 2 14 CH 3 SOH CML 7. 4 204. 9 130. 1 12 NH 2 CO 2 H MG-H 23. 7 229. 2 114. 3 14 NH 2 CH(CO 2 H)CH 2 CH=CH 2 250. 4 22 NH 2 CH(CO 2 H)CH 2 CH=CH 2 Pent 16. 5 379. 3

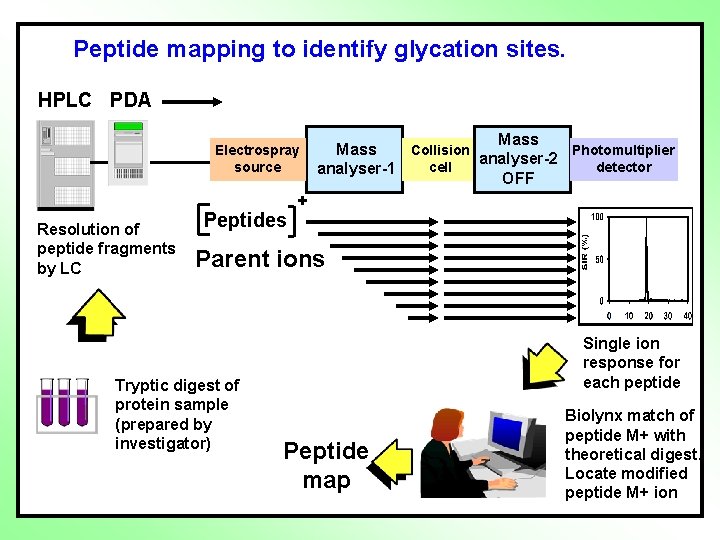
Peptide mapping to identify sites glycation of protein sites. modification Mass spectrometer HPLC PDA Electrospray source Resolution of peptide fragments by LC Peptides Mass Photomultiplier Collision analyser-2 detector cell analyser-1 OFF + Parent ions Tryptic digest of protein sample (prepared by investigator) Single ion response for each peptide Peptide map Biolynx match of peptide M+ with theoretical digest. Locate modified peptide M+ ion

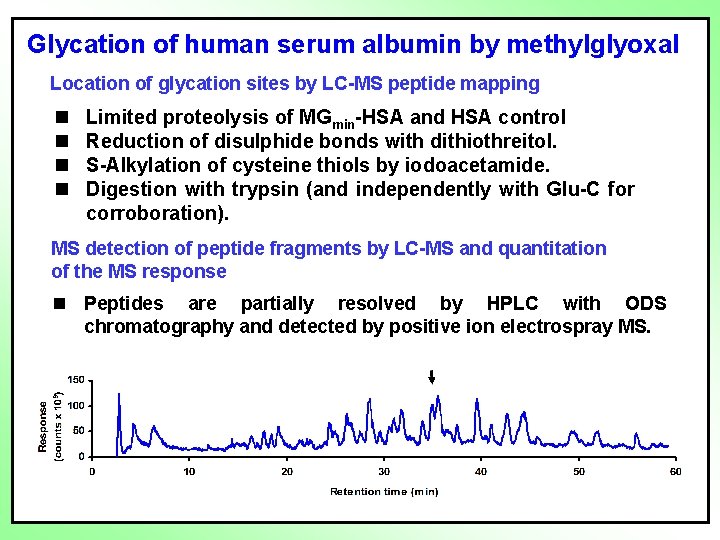
Glycation of human serum albumin by methylglyoxal Location of glycation sites by LC-MS peptide mapping n n Limited proteolysis of MGmin-HSA and HSA control Reduction of disulphide bonds with dithiothreitol. S-Alkylation of cysteine thiols by iodoacetamide. Digestion with trypsin (and independently with Glu-C for corroboration). MS detection of peptide fragments by LC-MS and quantitation of the MS response n Peptides are partially resolved by HPLC with ODS chromatography and detected by positive ion electrospray MS.

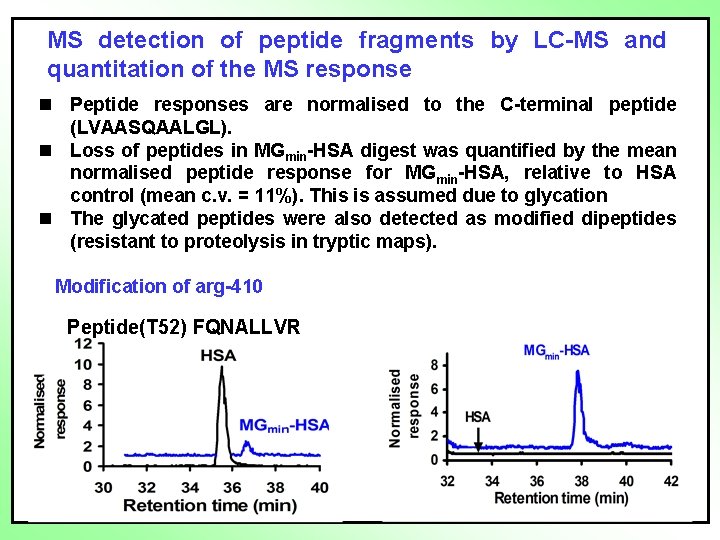
MS detection of peptide fragments by LC-MS and quantitation of the MS response n Peptide responses are normalised to the C-terminal peptide (LVAASQAALGL). n Loss of peptides in MGmin-HSA digest was quantified by the mean normalised peptide response for MGmin-HSA, relative to HSA control (mean c. v. = 11%). This is assumed due to glycation n The glycated peptides were also detected as modified dipeptides (resistant to proteolysis in tryptic maps). Modification of arg-410 Peptide(T 52) FQNALLVR

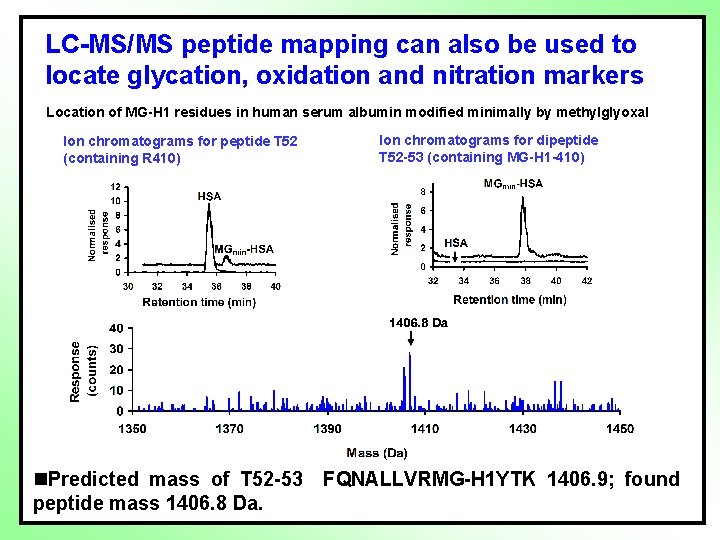
LC-MS/MS peptide mapping can also be used to locate glycation, oxidation and nitration markers Location of MG-H 1 residues in human serum albumin modified minimally by methylglyoxal Ion chromatograms for peptide T 52 (containing R 410) Ion chromatograms for dipeptide T 52 -53 (containing MG-H 1 -410) n. Predicted mass of T 52 -53 FQNALLVRMG-H 1 YTK 1406. 9; found peptide mass 1406. 8 Da.

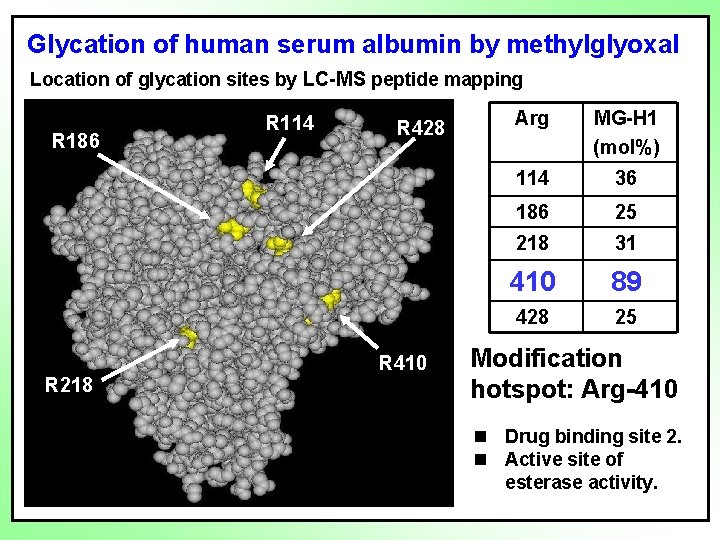
Glycation of human serum albumin by methylglyoxal Location of glycation sites by LC-MS peptide mapping R 186 R 218 R 114 R 428 R 410 Arg MG-H 1 (mol%) 114 36 186 25 218 31 410 89 428 25 Modification hotspot: Arg-410 n Drug binding site 2. n Active site of esterase activity.
 Il rombo ha tutti i lati uguali
Il rombo ha tutti i lati uguali Fluorescence microscopy uses
Fluorescence microscopy uses Laser confocal microscopy
Laser confocal microscopy Limitations of the beer lambert law
Limitations of the beer lambert law Atomic absorption spectrophotometer ppt
Atomic absorption spectrophotometer ppt Chemiluminescence vs fluorescence
Chemiluminescence vs fluorescence Principles of fluorescence spectroscopy
Principles of fluorescence spectroscopy F=p/2
F=p/2 Cold vapor atomic fluorescence spectrometry
Cold vapor atomic fluorescence spectrometry Fluorescence recovery after photobleaching
Fluorescence recovery after photobleaching Rfu fluorescence
Rfu fluorescence Fluorescence activated cell sorting
Fluorescence activated cell sorting Principle of fluorescence spectroscopy
Principle of fluorescence spectroscopy Fluorescence-activated cell sorting (facs)
Fluorescence-activated cell sorting (facs) Jablonski diagram fluorescence
Jablonski diagram fluorescence Light sources for fluorescence microscopy
Light sources for fluorescence microscopy Time to amplitude converter tac
Time to amplitude converter tac Compound microscope use
Compound microscope use