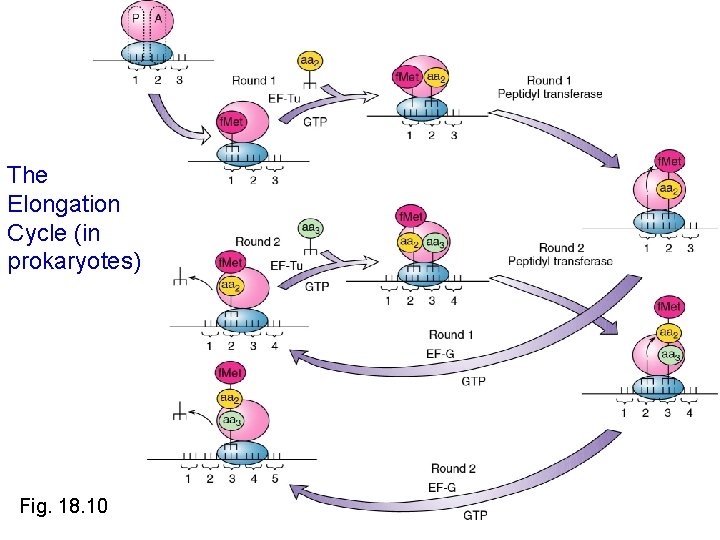
The Elongation Cycle in prokaryotes Fig 18 10


- Slides: 21

The Elongation Cycle (in prokaryotes) Fig. 18. 10

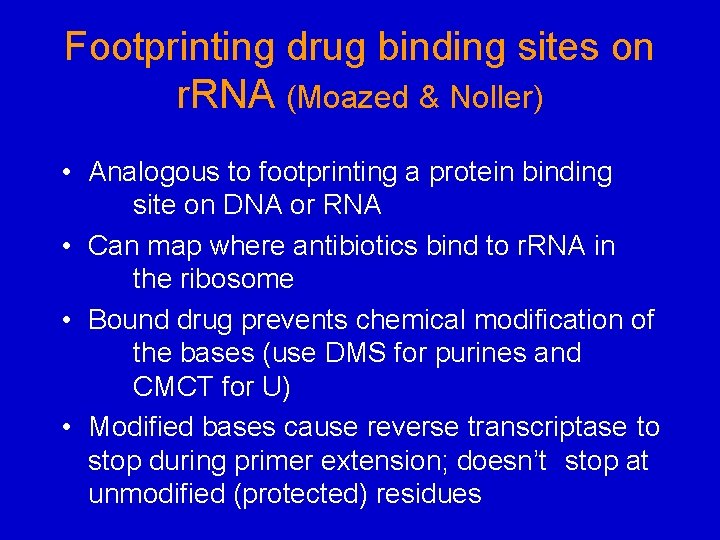
Footprinting drug binding sites on r. RNA (Moazed & Noller) • Analogous to footprinting a protein binding site on DNA or RNA • Can map where antibiotics bind to r. RNA in the ribosome • Bound drug prevents chemical modification of the bases (use DMS for purines and CMCT for U) • Modified bases cause reverse transcriptase to stop during primer extension; doesn’t stop at unmodified (protected) residues

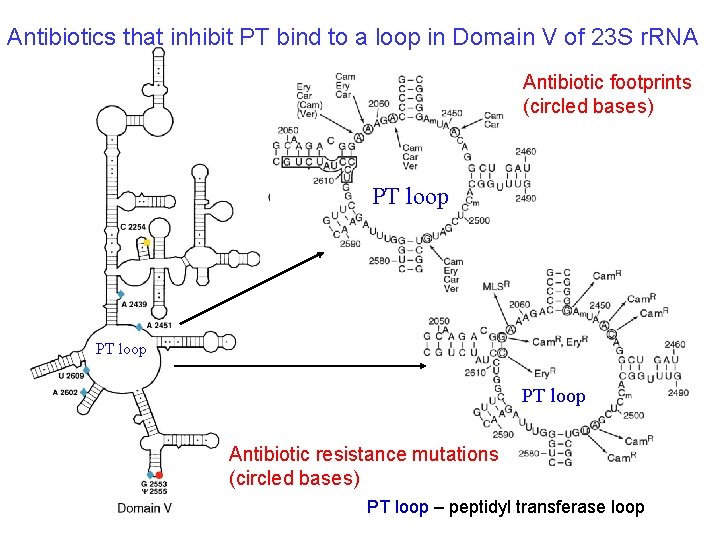
Antibiotics that inhibit PT bind to a loop in Domain V of 23 S r. RNA Antibiotic footprints (circled bases) PT loop Antibiotic resistance mutations (circled bases) PT loop – peptidyl transferase loop

Locating the peptidyl transferase on the large ribosomal subunit

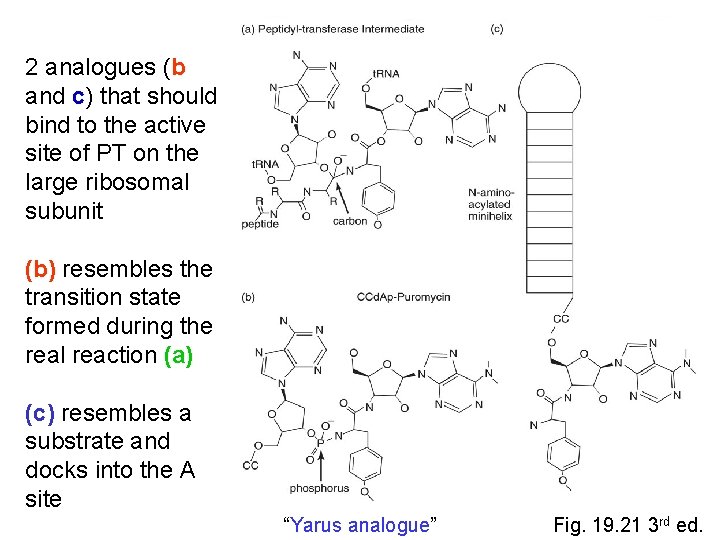
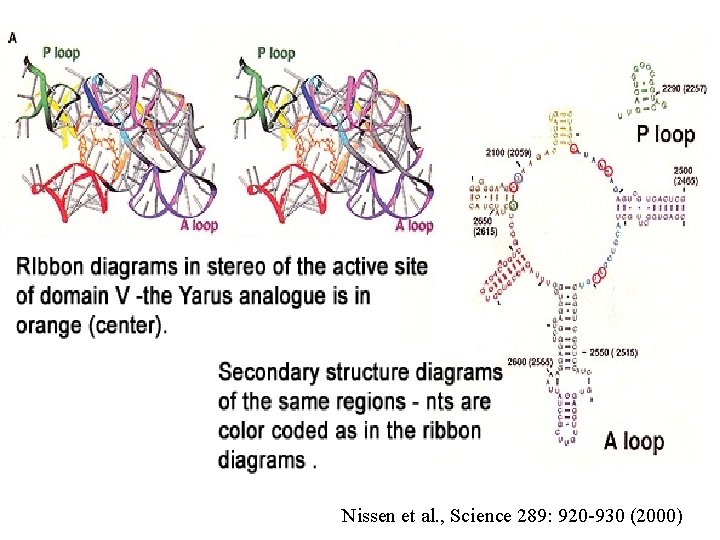
2 analogues (b and c) that should bind to the active site of PT on the large ribosomal subunit (b) resembles the transition state formed during the real reaction (a) (c) resembles a substrate and docks into the A site “Yarus analogue” Fig. 19. 21 3 rd ed.

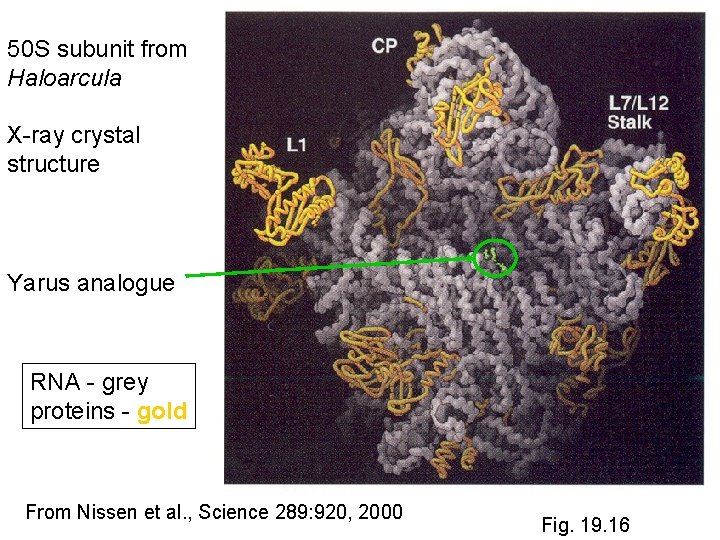
50 S subunit from Haloarcula X-ray crystal structure Yarus analogue RNA - grey proteins - gold From Nissen et al. , Science 289: 920, 2000 Fig. 19. 16

Nissen et al. , Science 289: 920 -930 (2000)

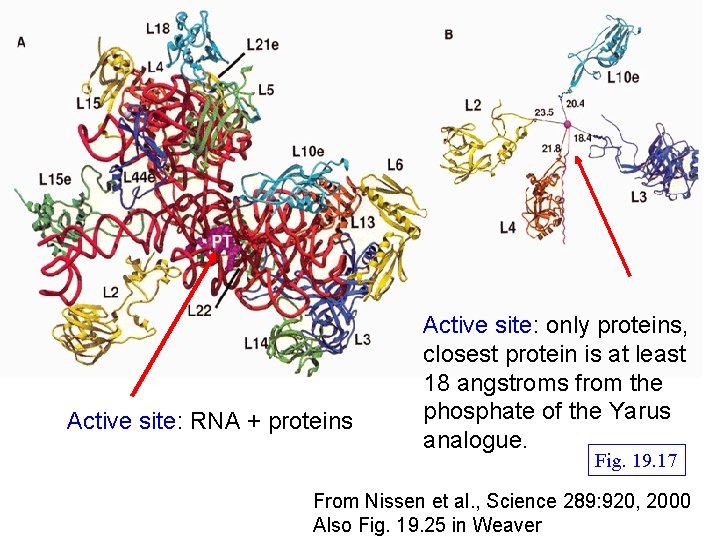
Active site: RNA + proteins Active site: only proteins, closest protein is at least 18 angstroms from the phosphate of the Yarus analogue. Fig. 19. 17 From Nissen et al. , Science 289: 920, 2000 Also Fig. 19. 25 in Weaver

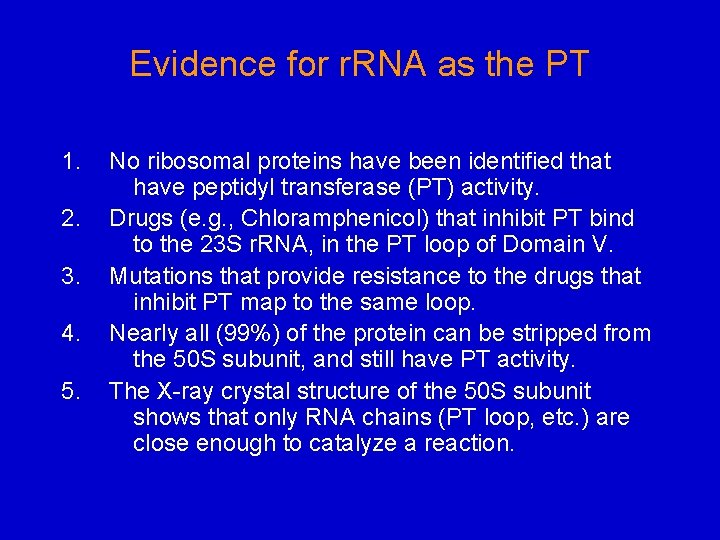
Evidence for r. RNA as the PT 1. 2. 3. 4. 5. No ribosomal proteins have been identified that have peptidyl transferase (PT) activity. Drugs (e. g. , Chloramphenicol) that inhibit PT bind to the 23 S r. RNA, in the PT loop of Domain V. Mutations that provide resistance to the drugs that inhibit PT map to the same loop. Nearly all (99%) of the protein can be stripped from the 50 S subunit, and still have PT activity. The X-ray crystal structure of the 50 S subunit shows that only RNA chains (PT loop, etc. ) are close enough to catalyze a reaction.

• Are there any potential deficiencies with this model or the data that support it? • How could it be made stronger?

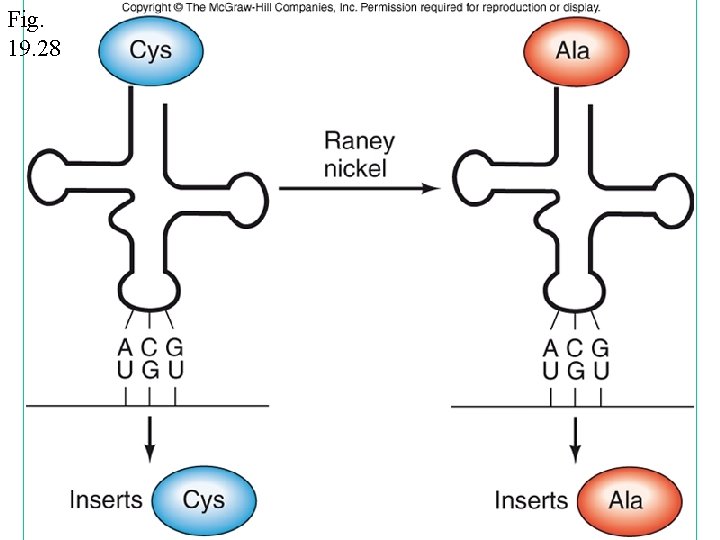
Fig. 19. 28

t. RNA Charging: The Second Genetic Code 1. t. RNA structure 2. the charging reaction 3. aminoacyl t. RNA synthetases and t. RNA recognition 4. proofreading mechanism

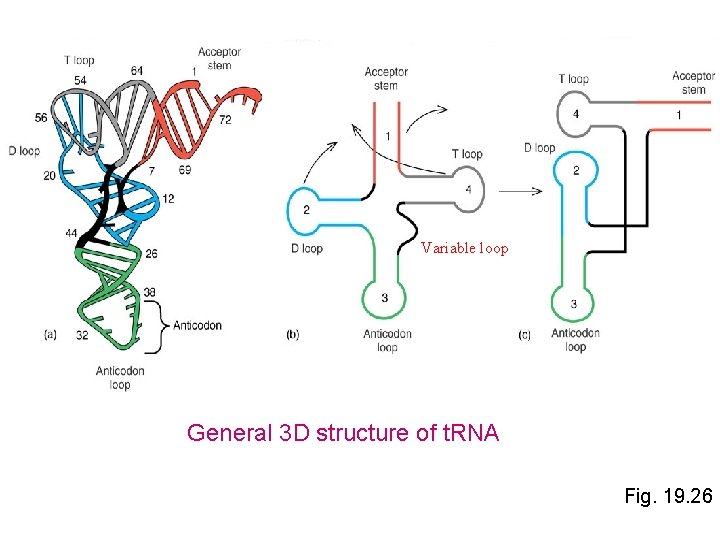
Variable loop General 3 D structure of t. RNA Fig. 19. 26

Fig. 19. 25

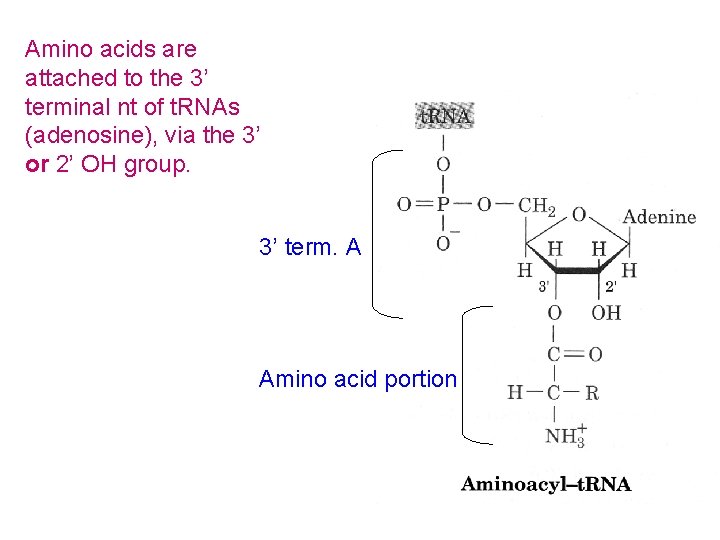
Amino acids are attached to the 3’ terminal nt of t. RNAs (adenosine), via the 3’ or 2’ OH group. 3’ term. A Amino acid portion

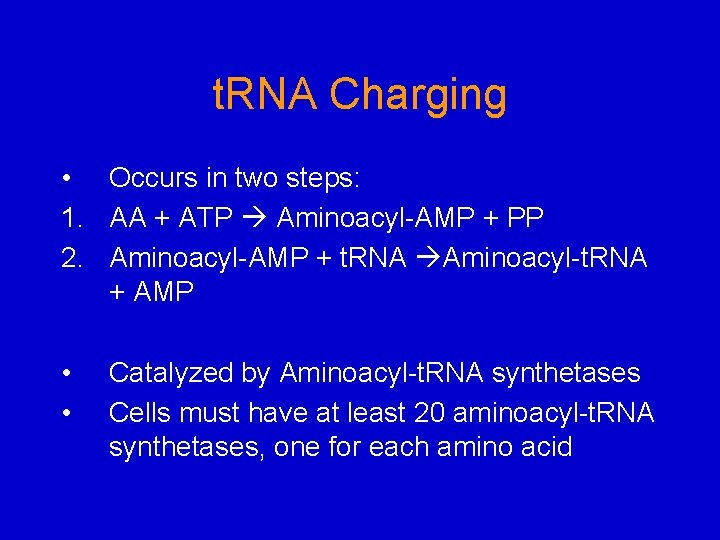
t. RNA Charging • Occurs in two steps: 1. AA + ATP Aminoacyl-AMP + PP 2. Aminoacyl-AMP + t. RNA Aminoacyl-t. RNA + AMP • • Catalyzed by Aminoacyl-t. RNA synthetases Cells must have at least 20 aminoacyl-t. RNA synthetases, one for each amino acid

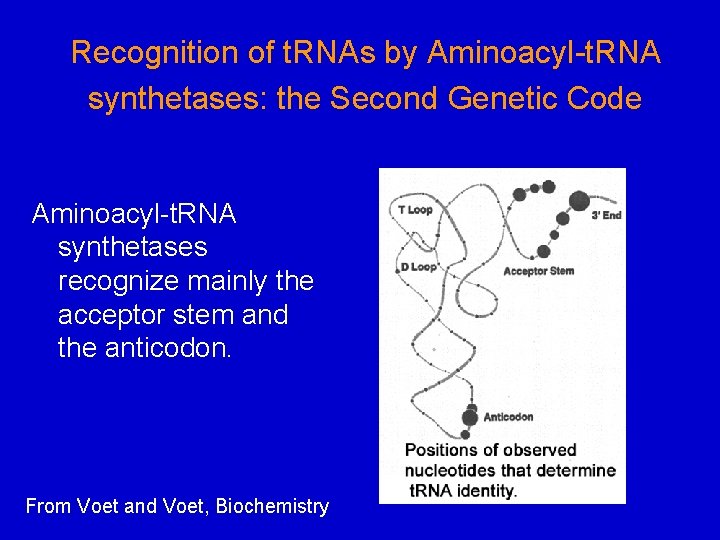
Recognition of t. RNAs by Aminoacyl-t. RNA synthetases: the Second Genetic Code Aminoacyl-t. RNA synthetases recognize mainly the acceptor stem and the anticodon. From Voet and Voet, Biochemistry

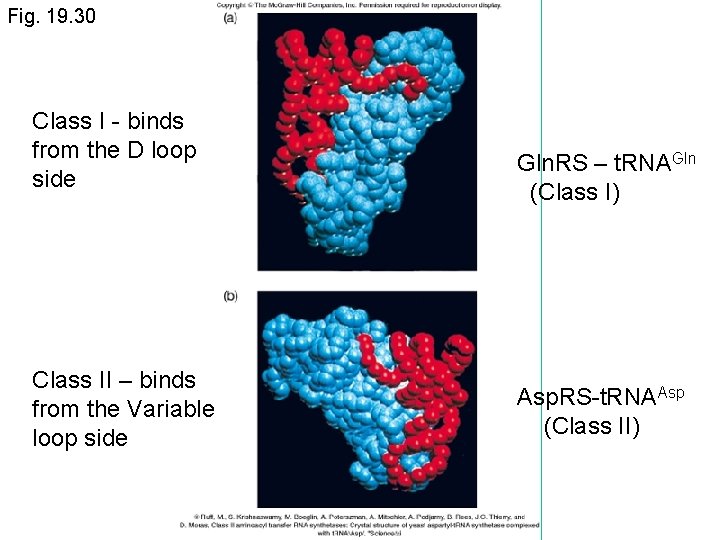
Aminoacyl-t. RNA synthetases (cont. ) • Diverse group of enzymes despite recognizing fairly similar substrates • Not well conserved, however there are 2 main classes – Class I (aminoacylate the 2’ OH) – Class II (aminoacylate the 3’ OH) • Each class has the same 10 members in all organisms • The classes bind t. RNA somewhat differently, but both bind to the acceptor stem and the anticodon loop

Fig. 19. 30 Class I - binds from the D loop side Class II – binds from the Variable loop side Gln. RS – t. RNAGln (Class I) Asp. RS-t. RNAAsp (Class II)

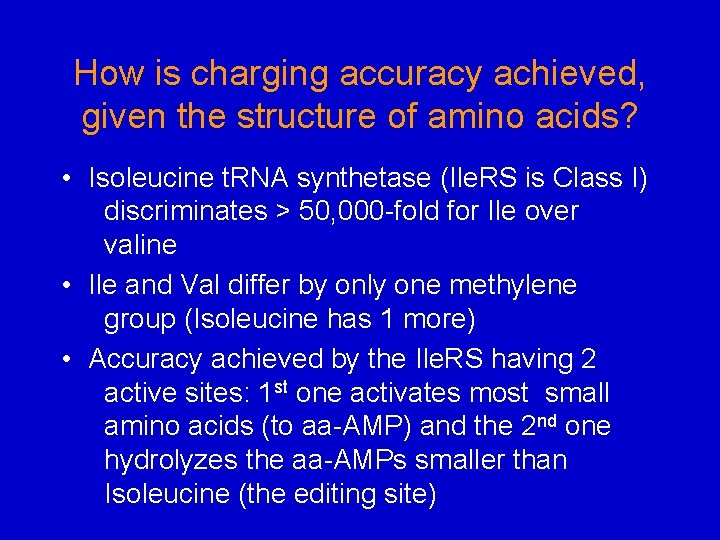
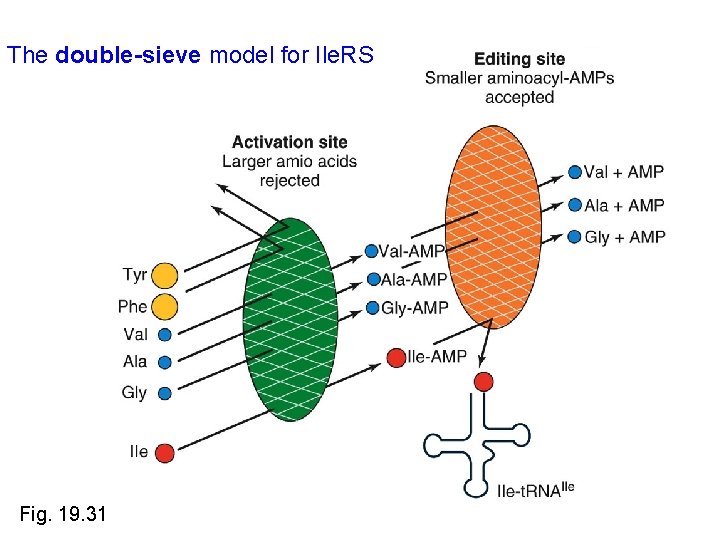
How is charging accuracy achieved, given the structure of amino acids? • Isoleucine t. RNA synthetase (Ile. RS is Class I) discriminates > 50, 000 -fold for Ile over valine • Ile and Val differ by only one methylene group (Isoleucine has 1 more) • Accuracy achieved by the Ile. RS having 2 active sites: 1 st one activates most small amino acids (to aa-AMP) and the 2 nd one hydrolyzes the aa-AMPs smaller than Isoleucine (the editing site)

The double-sieve model for Ile. RS Fig. 19. 31