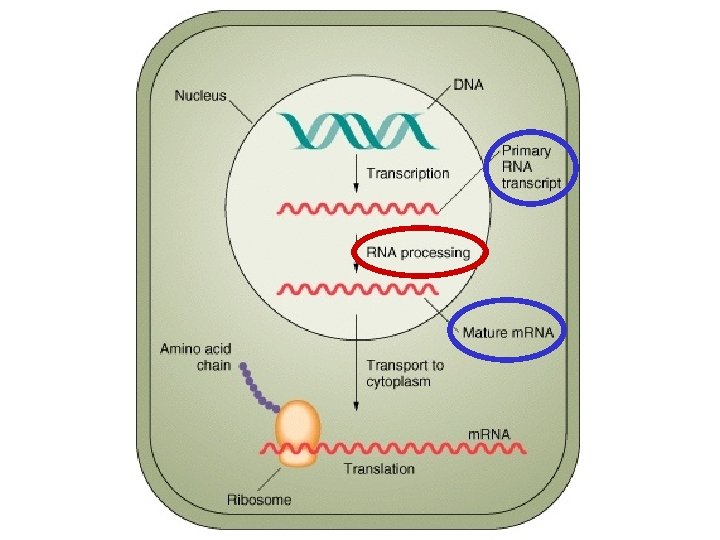
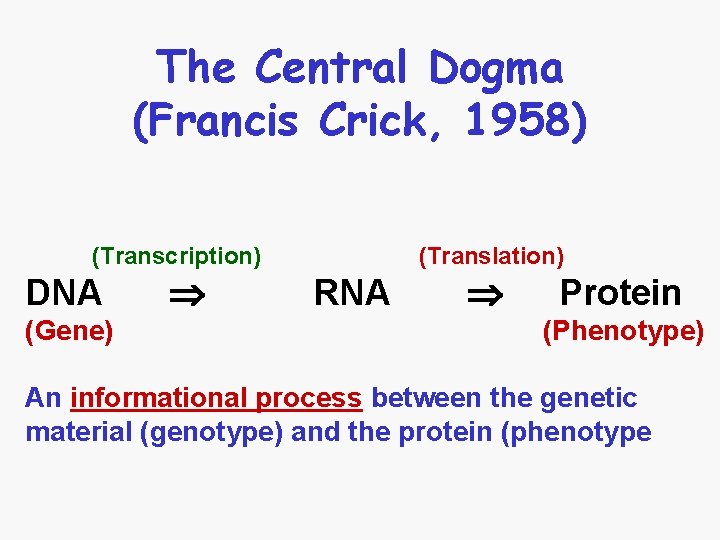
The Central Dogma Francis Crick 1958 Transcription DNA




- Slides: 25

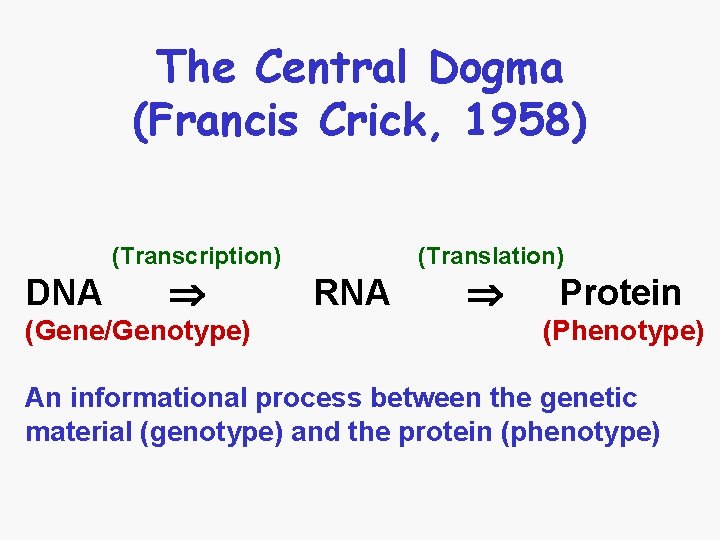
The Central Dogma (Francis Crick, 1958) (Transcription) DNA (Gene/Genotype) (Translation) RNA Protein (Phenotype) An informational process between the genetic material (genotype) and the protein (phenotype)

Classes of RNA for Transcription and Translation • Informational RNA (intermediate in the process of decoding genes into polypeptides) – Messenger RNA (m. RNA) • Functional RNAs (never translated into proteins, serve other roles) – Transfer RNAs (t. RNA) • Transport amino acids to m. RNA and new protein – Ribosomal RNAs (r. RNA) • Combine with an array of proteins to form ribosomes; platform for protein synthesis – Small nuclear RNAs (sn. RNA) • Take part in the splicing of primary transcripts in eukaryotes – Small cytoplasmic RNAs (sc. RNA) • Direct protein traffic in eukaryotic cells – Micro RNAs (mi. RNA) • Inhibits translation and induces degradation of complementary m. RNA

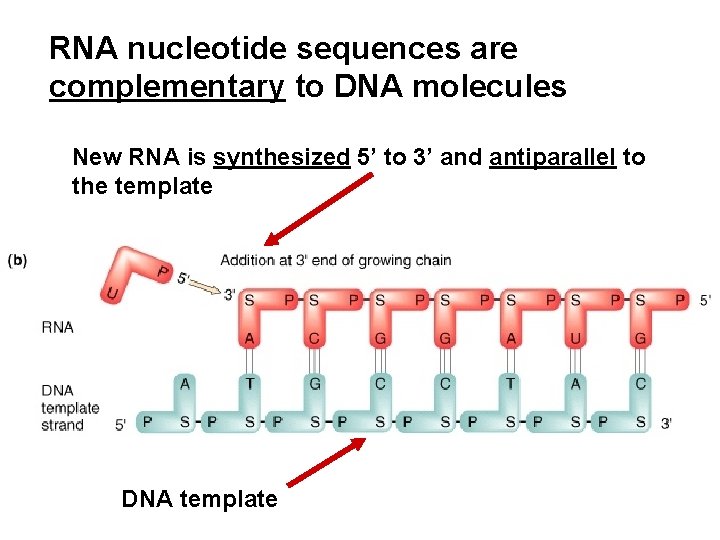
RNA nucleotide sequences are complementary to DNA molecules New RNA is synthesized 5’ to 3’ and antiparallel to the template DNA template

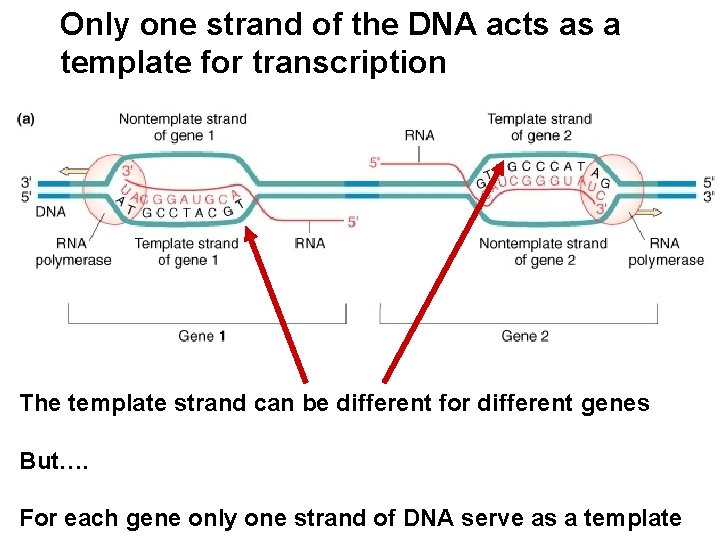
Only one strand of the DNA acts as a template for transcription The template strand can be different for different genes But…. For each gene only one strand of DNA serve as a template

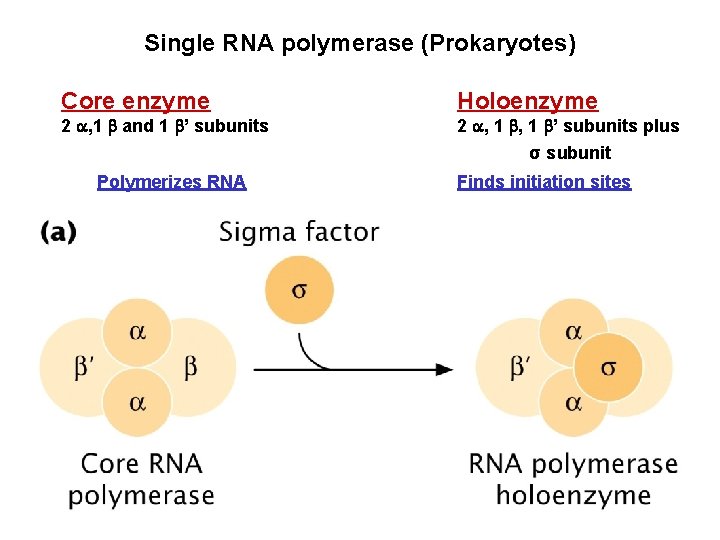
Single RNA polymerase (Prokaryotes) Core enzyme 2 , 1 and 1 ’ subunits Holoenzyme 2 , 1 ’ subunits plus σ subunit Polymerizes RNA Finds initiation sites

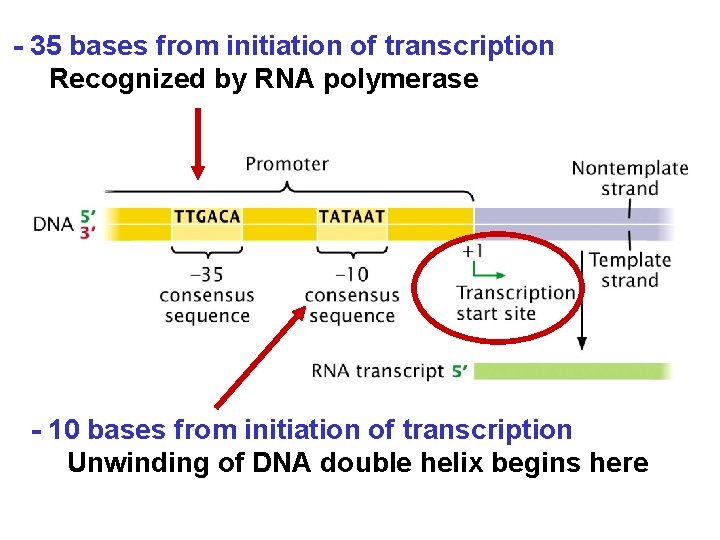
- 35 bases from initiation of transcription Recognized by RNA polymerase - 10 bases from initiation of transcription Unwinding of DNA double helix begins here

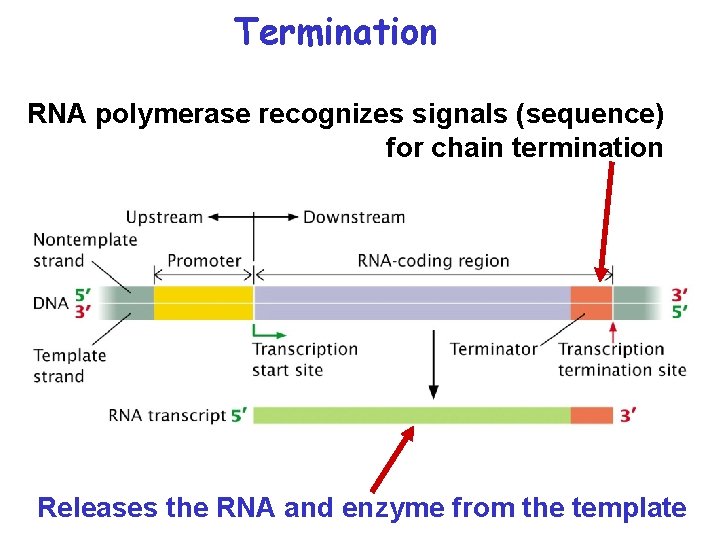
Termination RNA polymerase recognizes signals (sequence) for chain termination Releases the RNA and enzyme from the template


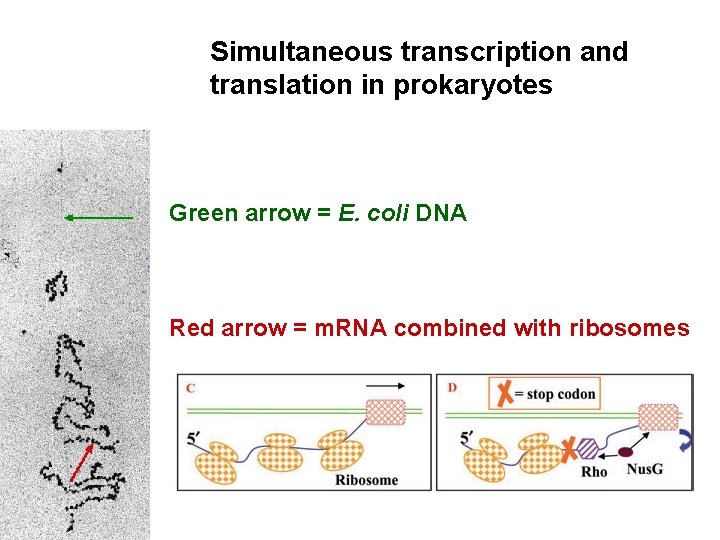
Simultaneous transcription and translation in prokaryotes Green arrow = E. coli DNA Red arrow = m. RNA combined with ribosomes


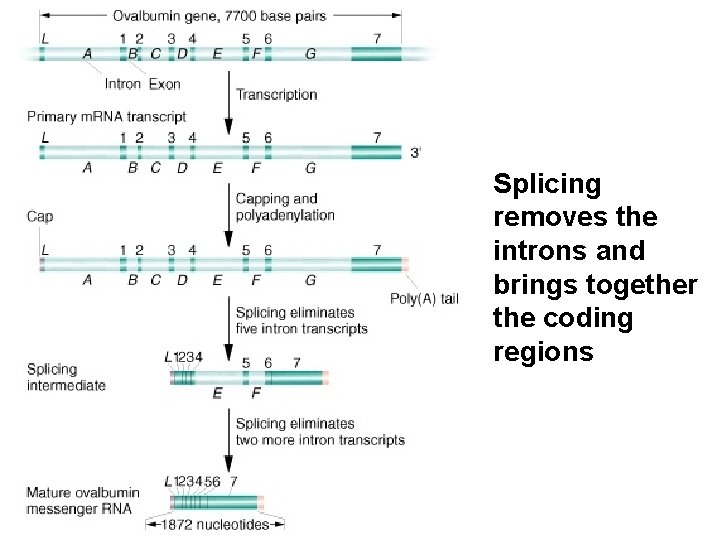
Splicing removes the introns and brings together the coding regions


The Central Dogma (Francis Crick, 1958) (Transcription) DNA (Gene) (Translation) RNA Protein (Phenotype) An informational process between the genetic material (genotype) and the protein (phenotype

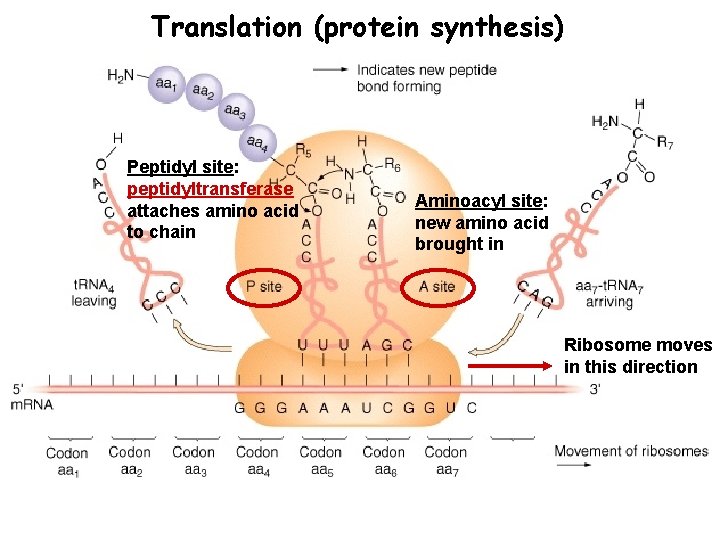
Translation (protein synthesis) Peptidyl site: peptidyltransferase attaches amino acid to chain Aminoacyl site: new amino acid brought in Ribosome moves in this direction

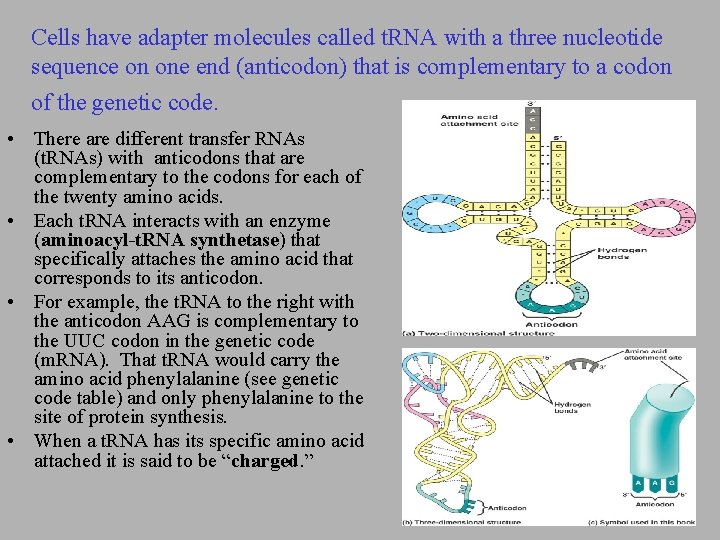
Cells have adapter molecules called t. RNA with a three nucleotide sequence on one end (anticodon) that is complementary to a codon of the genetic code. • There are different transfer RNAs (t. RNAs) with anticodons that are complementary to the codons for each of the twenty amino acids. • Each t. RNA interacts with an enzyme (aminoacyl-t. RNA synthetase) that specifically attaches the amino acid that corresponds to its anticodon. • For example, the t. RNA to the right with the anticodon AAG is complementary to the UUC codon in the genetic code (m. RNA). That t. RNA would carry the amino acid phenylalanine (see genetic code table) and only phenylalanine to the site of protein synthesis. • When a t. RNA has its specific amino acid attached it is said to be “charged. ”


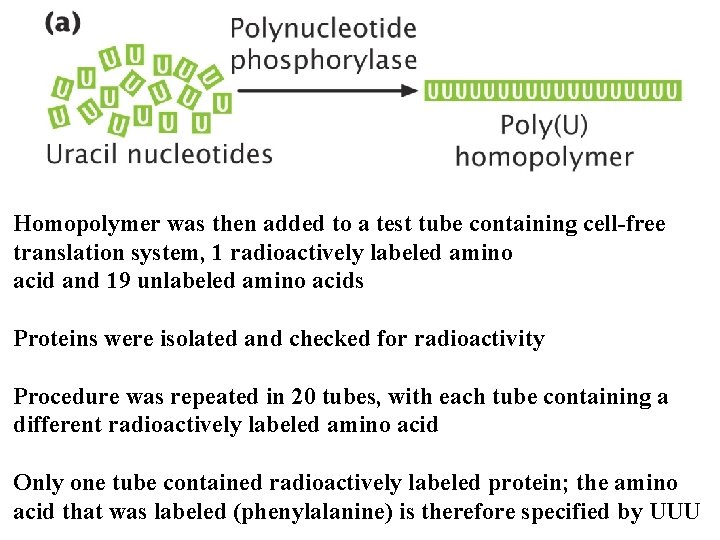
Homopolymer was then added to a test tube containing cell-free translation system, 1 radioactively labeled amino acid and 19 unlabeled amino acids Proteins were isolated and checked for radioactivity Procedure was repeated in 20 tubes, with each tube containing a different radioactively labeled amino acid Only one tube contained radioactively labeled protein; the amino acid that was labeled (phenylalanine) is therefore specified by UUU

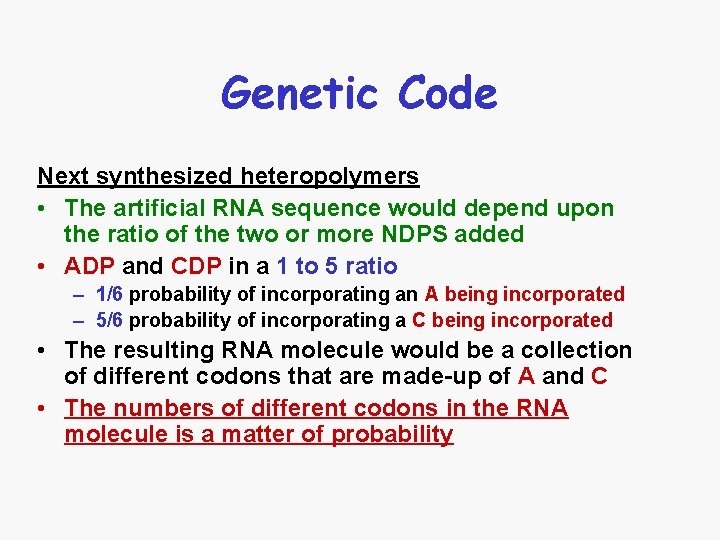
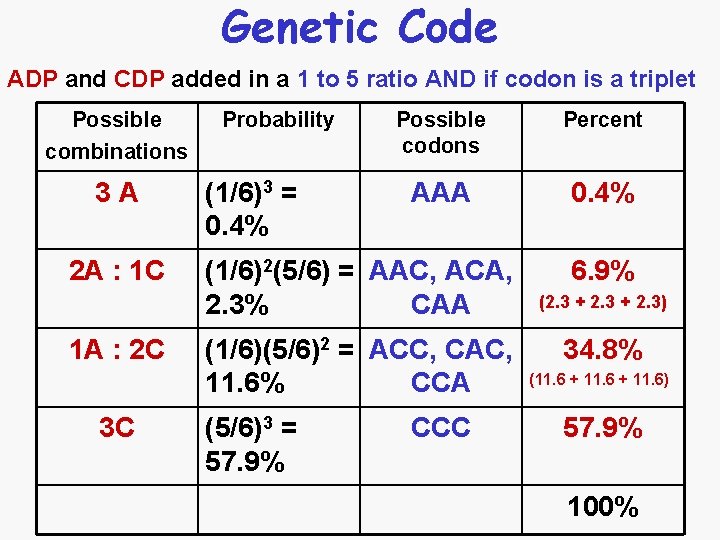
Genetic Code Next synthesized heteropolymers • The artificial RNA sequence would depend upon the ratio of the two or more NDPS added • ADP and CDP in a 1 to 5 ratio – 1/6 probability of incorporating an A being incorporated – 5/6 probability of incorporating a C being incorporated • The resulting RNA molecule would be a collection of different codons that are made-up of A and C • The numbers of different codons in the RNA molecule is a matter of probability

Genetic Code ADP and CDP added in a 1 to 5 ratio AND if codon is a triplet Possible combinations 3 A 2 A : 1 C 1 A : 2 C 3 C Probability (1/6)3 = 0. 4% Possible codons Percent AAA 0. 4% (1/6)2(5/6) = AAC, ACA, 2. 3% CAA (1/6)(5/6)2 = ACC, CAC, 11. 6% CCA (5/6)3 = 57. 9% CCC 6. 9% (2. 3 + 2. 3) 34. 8% (11. 6 + 11. 6) 57. 9% 100%

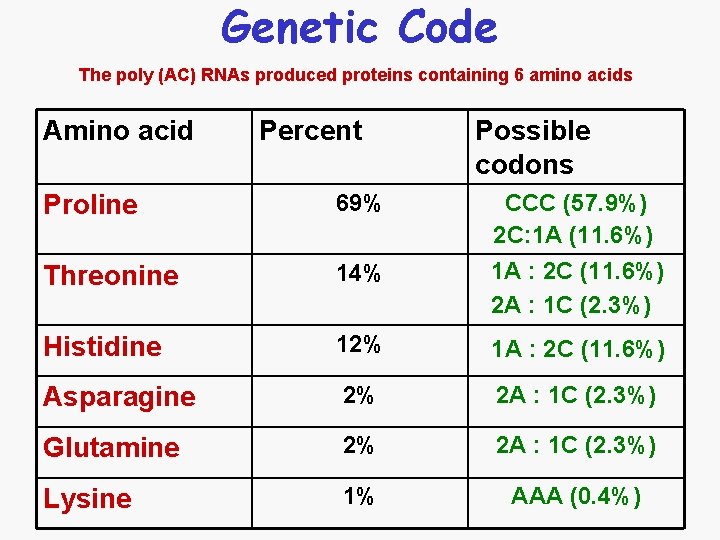
Genetic Code The poly (AC) RNAs produced proteins containing 6 amino acids Amino acid Percent Possible codons Proline 69% CCC (57. 9%) 2 C: 1 A (11. 6%) 1 A : 2 C (11. 6%) 2 A : 1 C (2. 3%) Threonine 14% Histidine 12% 1 A : 2 C (11. 6%) Asparagine 2% 2 A : 1 C (2. 3%) Glutamine 2% 2 A : 1 C (2. 3%) Lysine 1% AAA (0. 4%)

Using the table below, can you translate this nucleotide sequence? 5’UUCGAUGCCCGGGGUCCUGAAAUUGUUCUAGA 3’ • The first step is to look for the AUG start codon. • Next, group the nucleotides into a reading frame of 3 nucleotides per codons and use the table to find the amino acid that corresponds to each codon. • Stop translating the m. RNA when you reach a stop codon. • Is this what you got? Met-Pro-Gly-Val-Leu-Lys-Leu-Phe-Stop


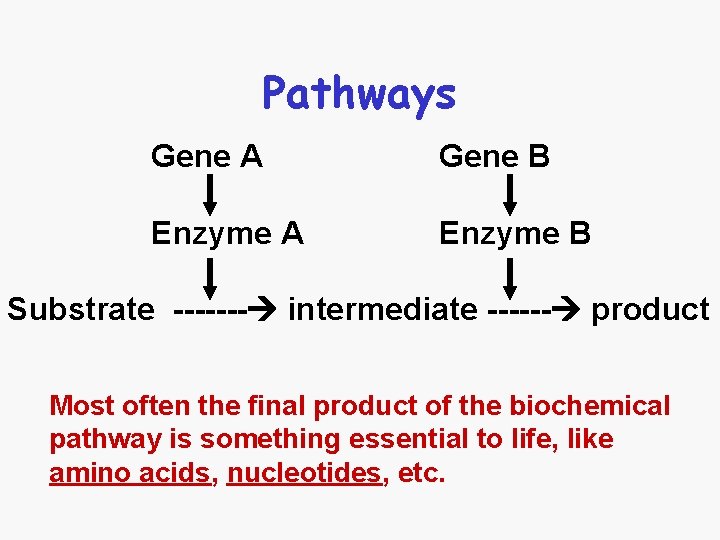
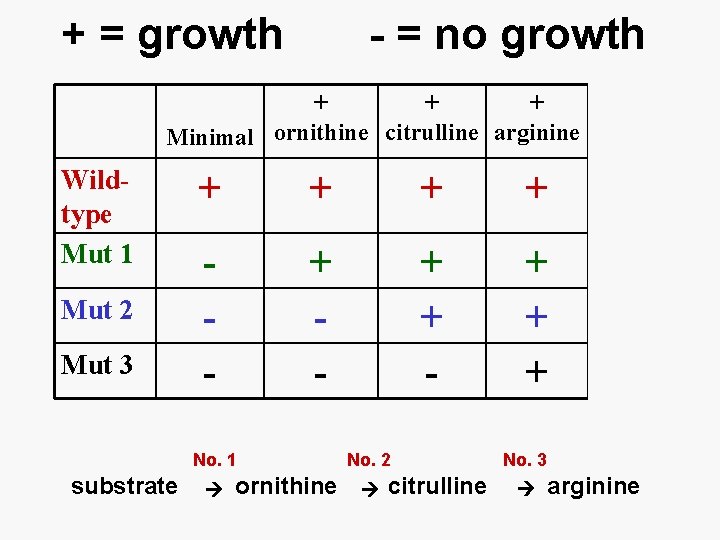
Pathways Gene A Gene B Enzyme A Enzyme B Substrate ------- intermediate ------ product Most often the final product of the biochemical pathway is something essential to life, like amino acids, nucleotides, etc.

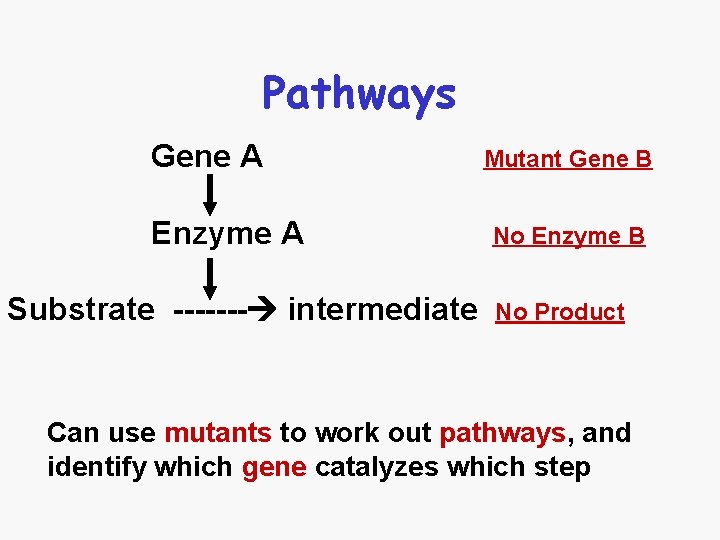
Pathways Gene A Enzyme A Substrate ------- intermediate Mutant Gene B No Enzyme B No Product Can use mutants to work out pathways, and identify which gene catalyzes which step

+ = growth - = no growth + + + Minimal ornithine citrulline arginine Wildtype Mut 1 Mut 2 Mut 3 + + 4 - + + + 3 2 1 No. 1 substrate No. +’s ornithine No. 2 citrulline No. 3 arginine