Technique 2 Each fragment is isolated with a

- Slides: 36


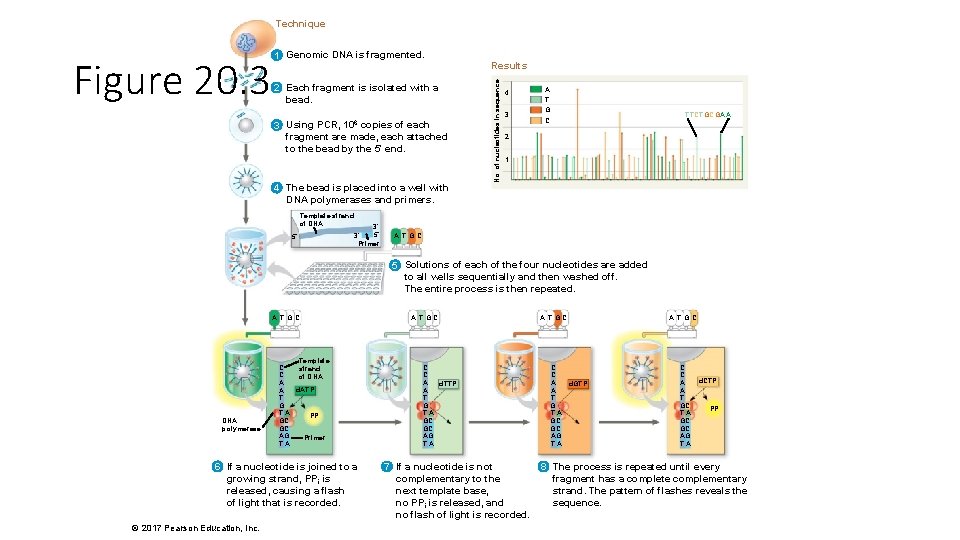
Technique 2 Each fragment is isolated with a bead. 3 Using PCR, 106 copies of each fragment are made, each attached to the bead by the 5′ end. 4 The bead is placed into a well with DNA polymerases and primers. Template strand of DNA 5′ 3′ Primer Results No. of nucleotides in sequence Figure 20. 3 1 Genomic DNA is fragmented. 4 3 A T G C TTCTGCGAA 2 1 A T GC 5 Solutions of each of the four nucleotides are added to all wells sequentially and then washed off. The entire process is then repeated. A TGC DNA polymerase Template C strand C of DNA A d. ATP A T G TA PPi GC GC AG Primer TA 6 If a nucleotide is joined to a growing strand, PPi is released, causing a flash of light that is recorded. © 2017 Pearson Education, Inc. A TGC C C A d. TTP A T G TA GC GC AG TA 7 If a nucleotide is not complementary to the next template base, no PPi is released, and no flash of light is recorded. A TGC C C A d. GTP A T G TA GC GC AG TA A TGC C C A A T GC TA GC GC AG TA d. CTP PPi 8 The process is repeated until every fragment has a complete complementary strand. The pattern of flashes reveals the sequence.

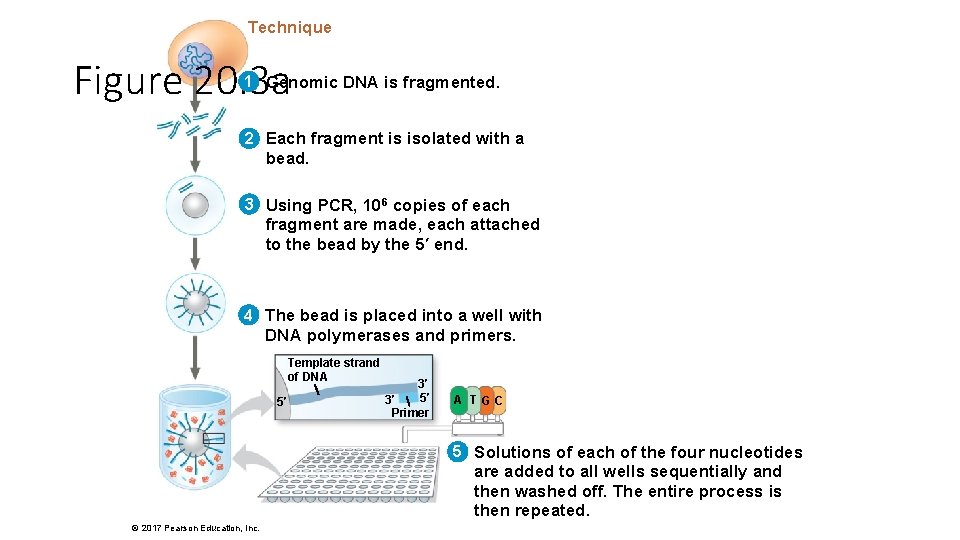
Technique Figure 20. 3 a 1 Genomic DNA is fragmented. 2 Each fragment is isolated with a bead. 3 Using PCR, 106 copies of each fragment are made, each attached to the bead by the 5′ end. 4 The bead is placed into a well with DNA polymerases and primers. Template strand of DNA 5′ 3′ Primer A T GC 5 Solutions of each of the four nucleotides are added to all wells sequentially and then washed off. The entire process is then repeated. © 2017 Pearson Education, Inc.

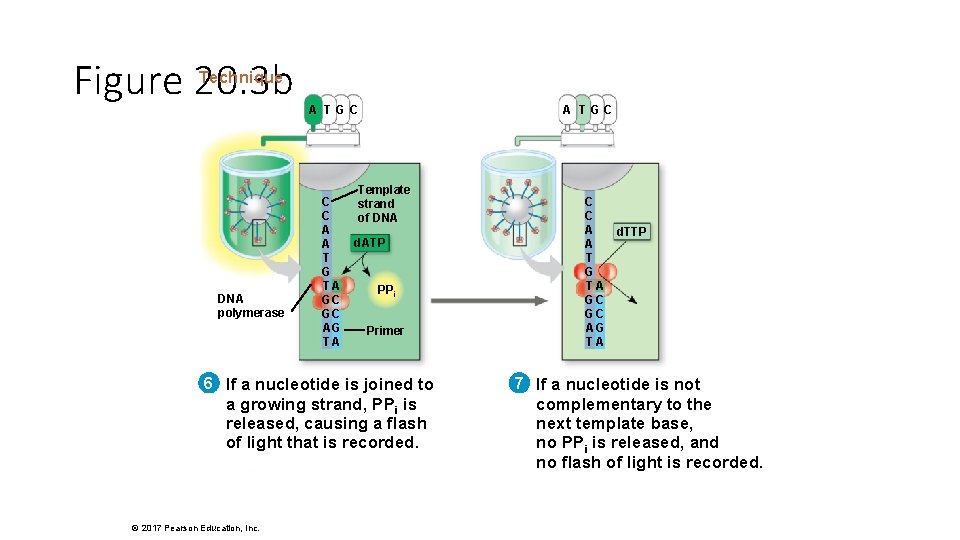
Figure 20. 3 b Technique DNA polymerase A TGC C C A A T G TA GC GC AG TA A TGC Template strand of DNA d. ATP PPi Primer 6 If a nucleotide is joined to a growing strand, PPi is released, causing a flash of light that is recorded. © 2017 Pearson Education, Inc. C C A d. TTP A T G TA GC GC AG TA 7 If a nucleotide is not complementary to the next template base, no PPi is released, and no flash of light is recorded.

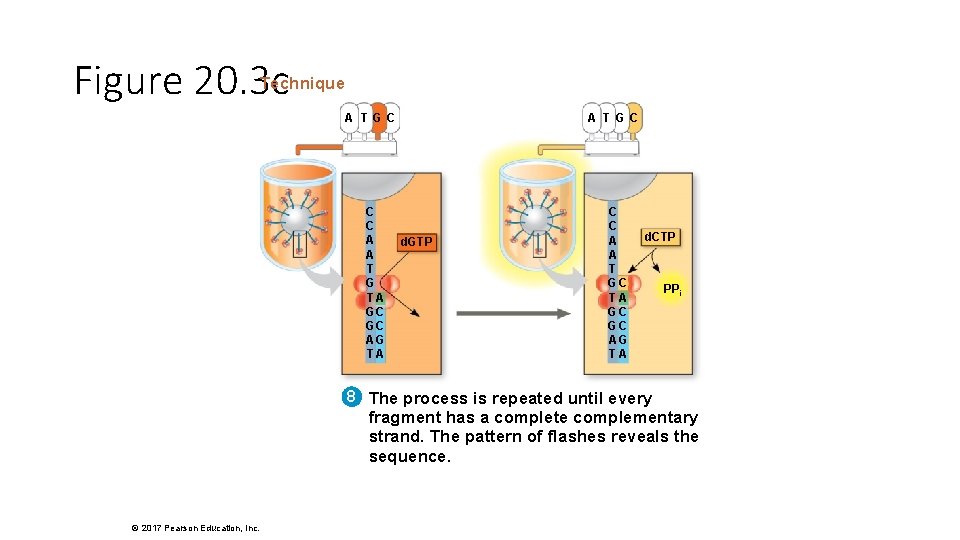
Figure 20. 3 c Technique A T GC C C A A T G TA GC GC AG TA A T GC d. GTP C C A A T GC TA GC GC AG TA d. CTP PPi 8 The process is repeated until every fragment has a complete complementary strand. The pattern of flashes reveals the sequence. © 2017 Pearson Education, Inc.

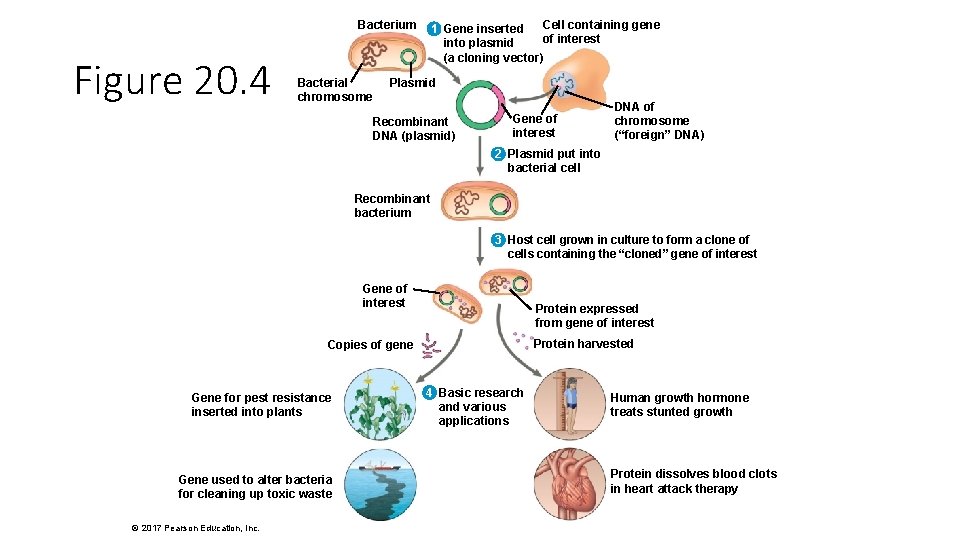
Bacterium Figure 20. 4 Bacterial chromosome Cell containing gene 1 Gene inserted of interest into plasmid (a cloning vector) Plasmid Recombinant DNA (plasmid) Gene of interest DNA of chromosome (“foreign” DNA) 2 Plasmid put into bacterial cell Recombinant bacterium 3 Host cell grown in culture to form a clone of cells containing the “cloned” gene of interest Gene of interest Protein expressed from gene of interest Protein harvested Copies of gene Gene for pest resistance inserted into plants Gene used to alter bacteria for cleaning up toxic waste © 2017 Pearson Education, Inc. 4 Basic research and various applications Human growth hormone treats stunted growth Protein dissolves blood clots in heart attack therapy

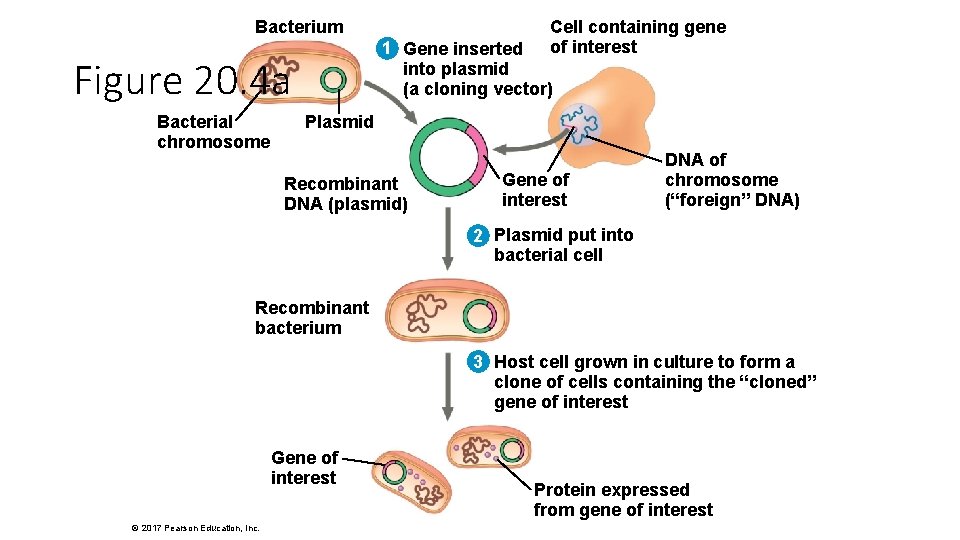
Bacterium 1 Gene inserted into plasmid (a cloning vector) Figure 20. 4 a Bacterial chromosome Cell containing gene of interest Plasmid Recombinant DNA (plasmid) Gene of interest DNA of chromosome (“foreign” DNA) 2 Plasmid put into bacterial cell Recombinant bacterium 3 Host cell grown in culture to form a clone of cells containing the “cloned” gene of interest Gene of interest © 2017 Pearson Education, Inc. Protein expressed from gene of interest

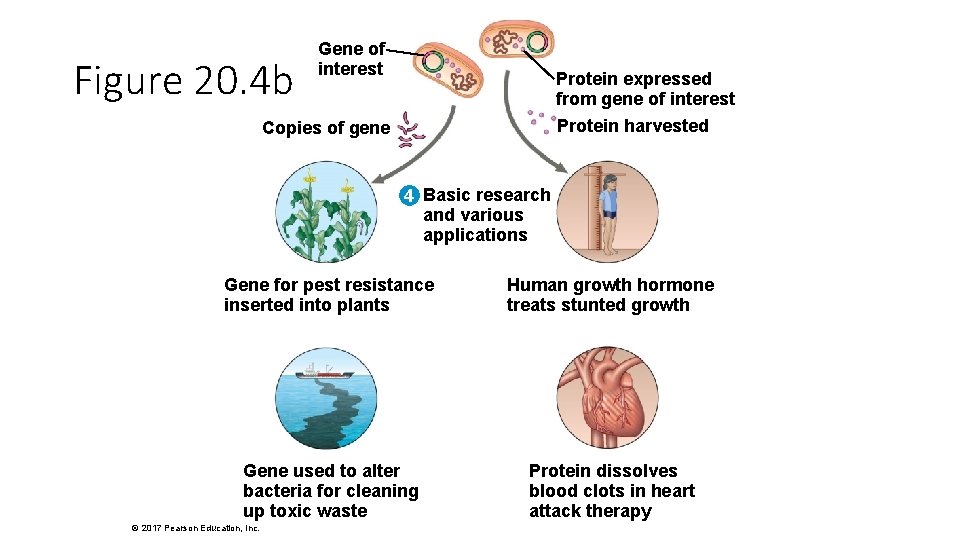
Figure 20. 4 b Gene of interest Protein expressed from gene of interest Protein harvested Copies of gene 4 Basic research and various applications Gene for pest resistance inserted into plants Human growth hormone treats stunted growth Gene used to alter bacteria for cleaning up toxic waste Protein dissolves blood clots in heart attack therapy © 2017 Pearson Education, Inc.

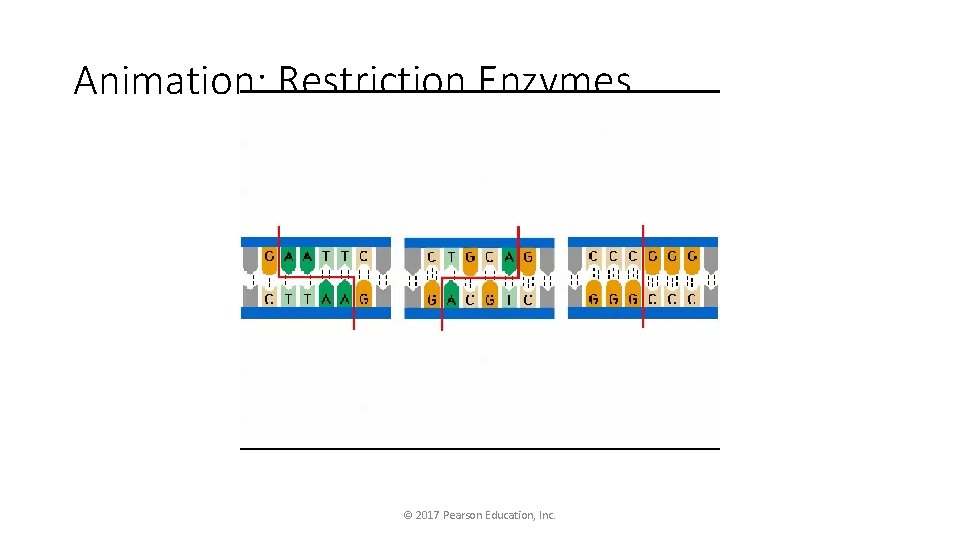
Animation: Restriction Enzymes © 2017 Pearson Education, Inc.

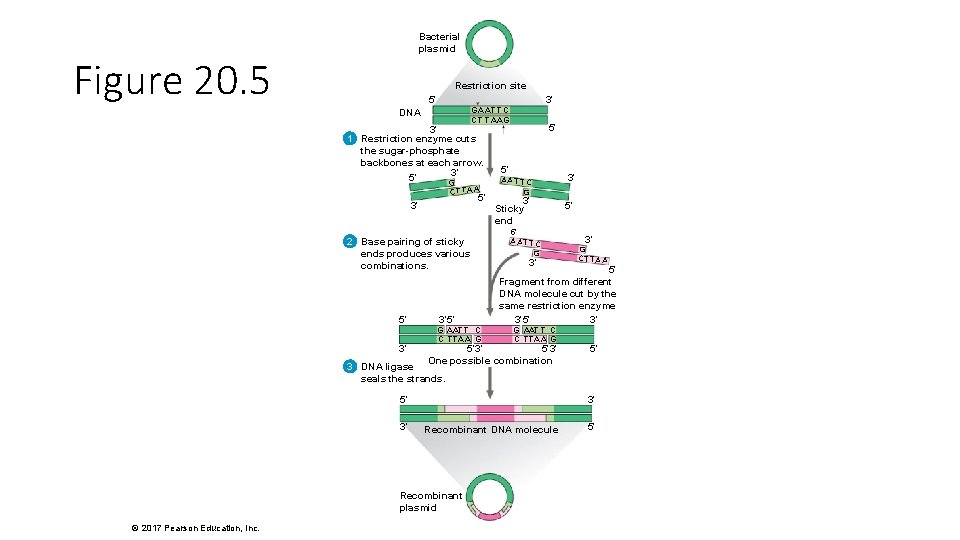
Bacterial plasmid Figure 20. 5 Restriction site 5′ 3′ GA AT T C CT T AAG DNA 3′ 1 Restriction enzyme cuts the sugar-phosphate backbones at each arrow. 3′ 5′ G CT TA A 5′ 3′ 2 Base pairing of sticky ends produces various combinations. 5′ 3′ 3′ 5′ G AAT T C C TTAA G 5′ 5′ A AT T C G 3′ Sticky end 5′ A AT T C G 3′ 3′ 5′ 3′ G CT TA A 5′ Fragment from different DNA molecule cut by the same restriction enzyme 3′ 5′ 3′ G AAT T C C TTAA G 5′ 3′ One possible combination 5′ 3 DNA ligase seals the strands. 5′ 3′ 3′ Recombinant DNA molecule Recombinant plasmid © 2017 Pearson Education, Inc. 5′

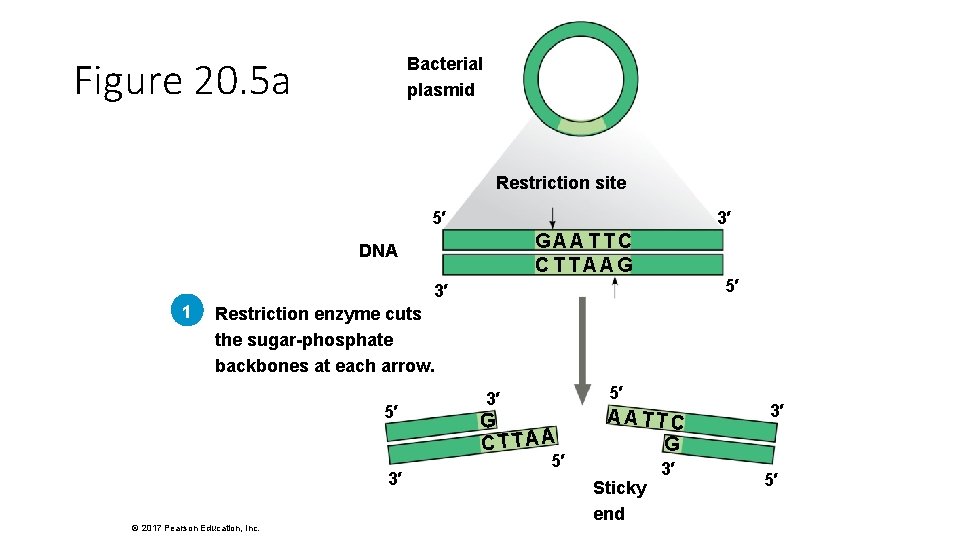
Figure 20. 5 a Bacterial plasmid Restriction site 5′ 3′ GAATTC CTTAAG DNA 5′ 3′ 1 Restriction enzyme cuts the sugar-phosphate backbones at each arrow. 5′ 3′ © 2017 Pearson Education, Inc. 5′ 3′ G CTTAA 5′ AATTC G Sticky end 3′ 3′ 5′

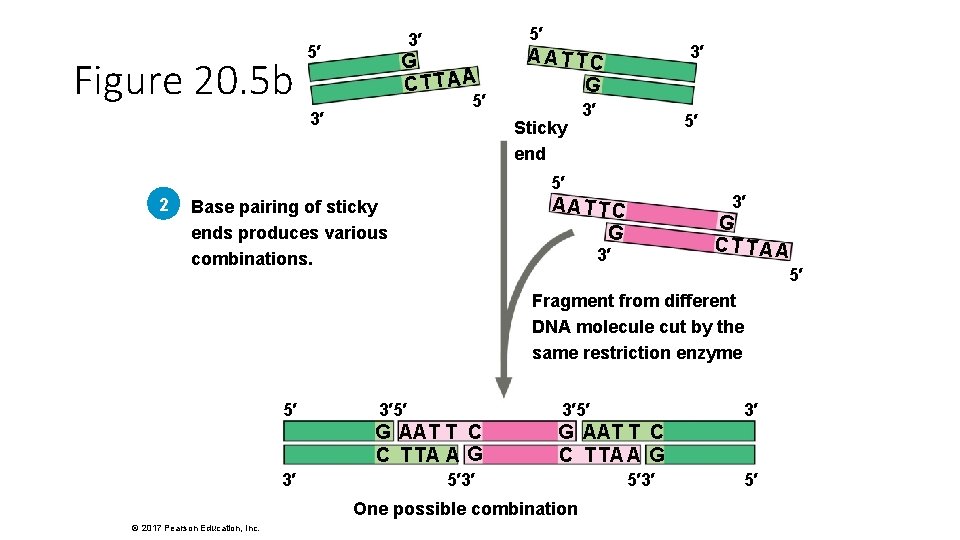
Figure 20. 5 b 5′ 3′ 5′ G CTTAA 5′ 3′ 3′ A AT TC G Sticky end 3′ 5′ 5′ 2 3′ AATTC G Base pairing of sticky ends produces various combinations. G CT TA A 3′ 5′ Fragment from different DNA molecule cut by the same restriction enzyme 5′ 3′ 3′ 5′ G AAT T C C T TA A G 5′ 3′ One possible combination © 2017 Pearson Education, Inc. 3′ 5′

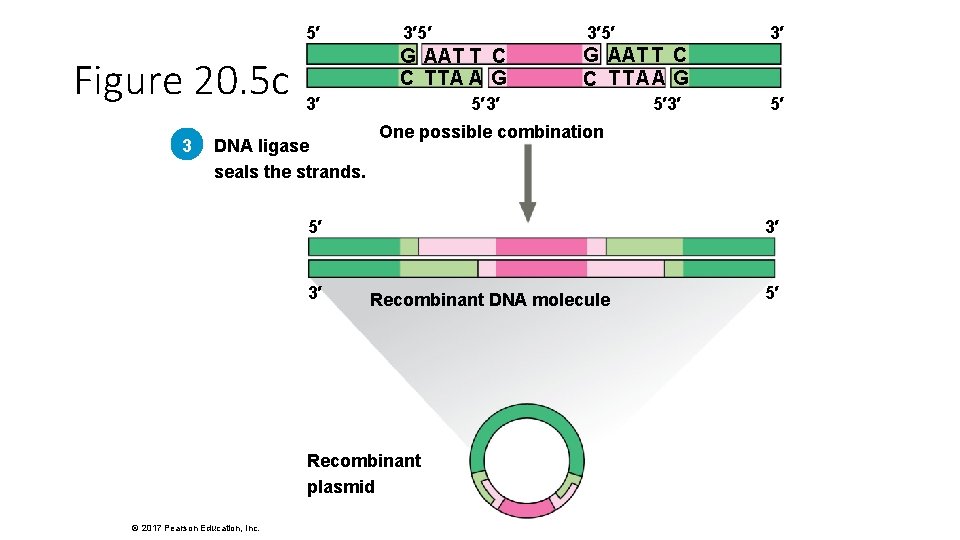
5′ Figure 20. 5 c 3 3′ 5′ G AAT T C C TTA A G G AAT T C C TTAA G 3′ DNA ligase seals the strands. 5′ 3′ Recombinant DNA molecule Recombinant plasmid © 2017 Pearson Education, Inc. 5′ 3′ One possible combination 5′ 3′ 3′ 5′

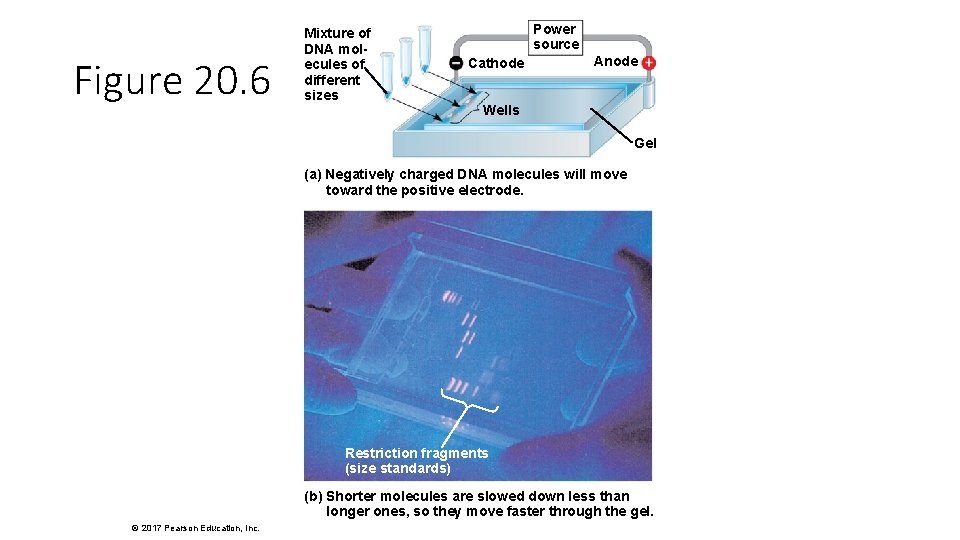
Figure 20. 6 Mixture of DNA molecules of different sizes Power source Cathode Anode Wells Gel (a) Negatively charged DNA molecules will move toward the positive electrode. Restriction fragments (size standards) (b) Shorter molecules are slowed down less than longer ones, so they move faster through the gel. © 2017 Pearson Education, Inc.

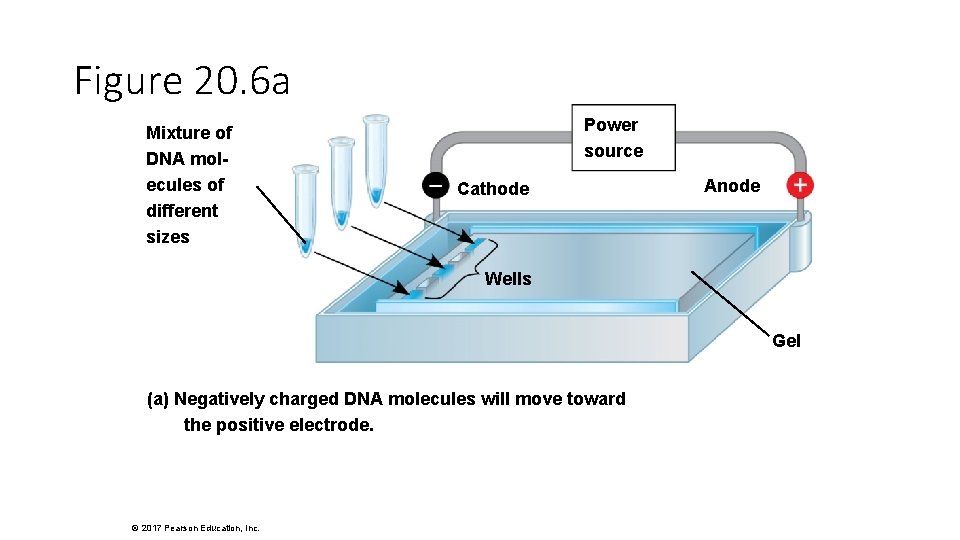
Figure 20. 6 a Mixture of DNA molecules of different sizes Power source Cathode Anode Wells Gel (a) Negatively charged DNA molecules will move toward the positive electrode. © 2017 Pearson Education, Inc.

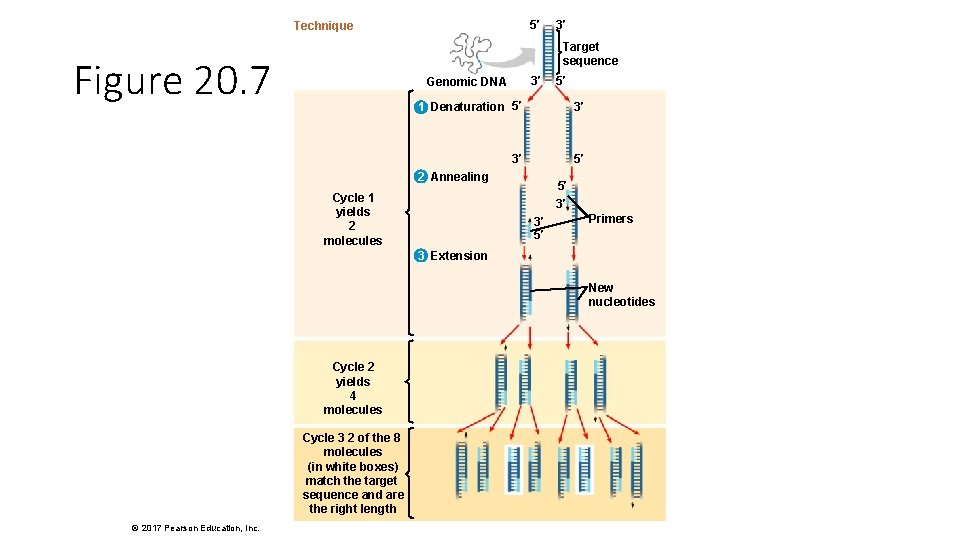
5′ Technique 3′ Target sequence Figure 20. 7 3′ Genomic DNA 5′ 1 Denaturation 5′ 3′ 3′ 5′ 2 Annealing Cycle 1 yields 2 molecules 5′ 3′ 3′ 5′ Primers 3 Extension New nucleotides Cycle 2 yields 4 molecules Cycle 3 2 of the 8 molecules (in white boxes) match the target sequence and are the right length © 2017 Pearson Education, Inc.

Figure 20. 7 a Technique 5′ 3′ Target sequence Genomic DNA © 2017 Pearson Education, Inc. 3′ 5′

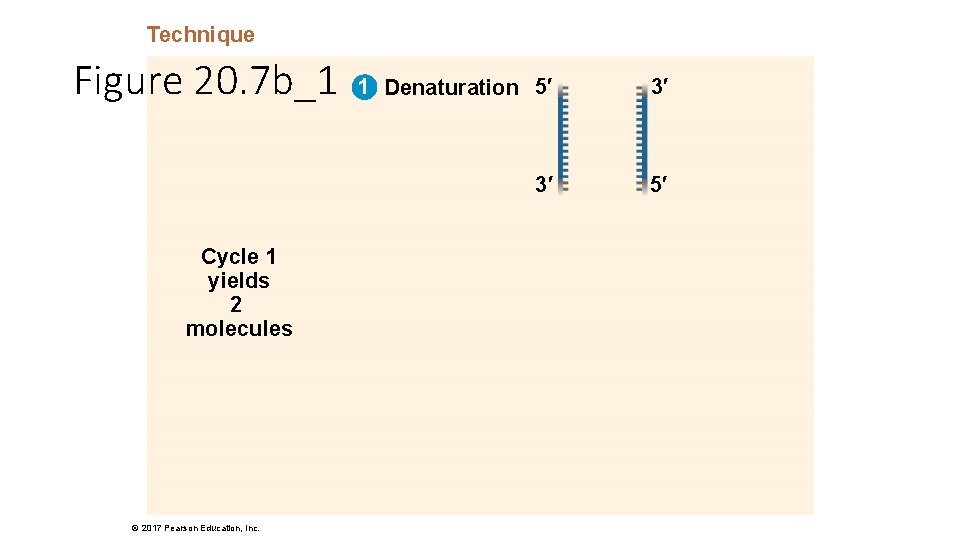
Technique Figure 20. 7 b_1 Cycle 1 yields 2 molecules © 2017 Pearson Education, Inc. 1 Denaturation 5′ 3′ 3′ 5′

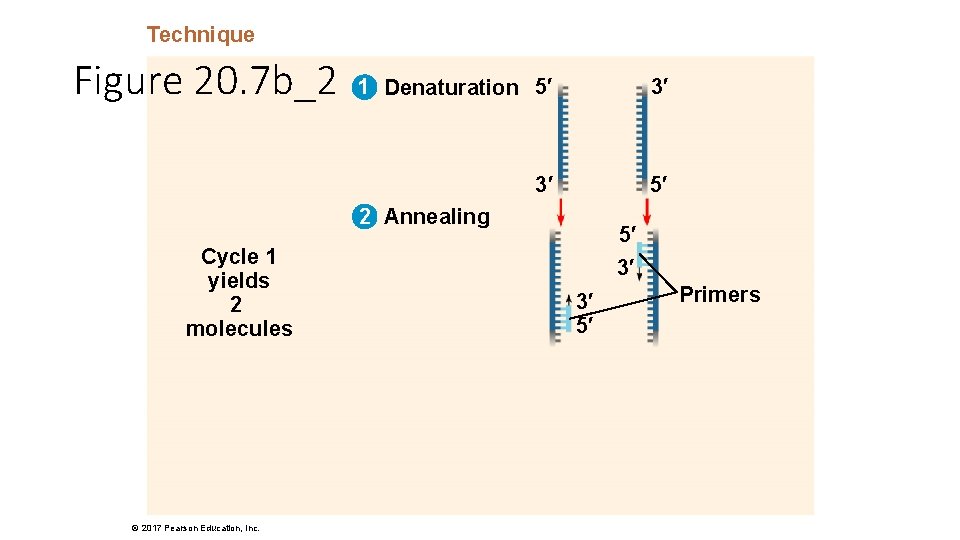
Technique Figure 20. 7 b_2 1 Denaturation 5′ 3′ 3′ 5′ 2 Annealing Cycle 1 yields 2 molecules © 2017 Pearson Education, Inc. 5′ 3′ 3′ 5′ Primers

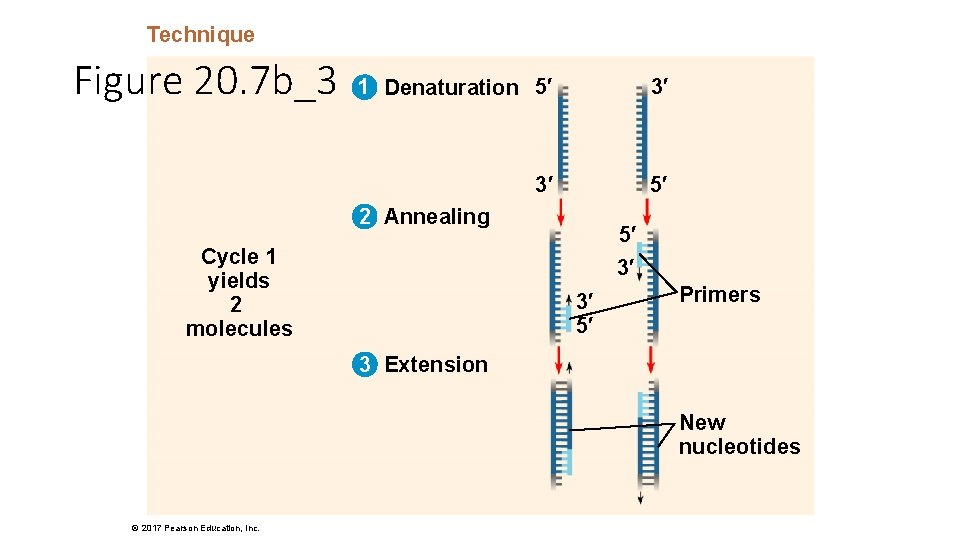
Technique Figure 20. 7 b_3 1 Denaturation 5′ 3′ 3′ 5′ 2 Annealing Cycle 1 yields 2 molecules 5′ 3′ 3′ 5′ Primers 3 Extension New nucleotides © 2017 Pearson Education, Inc.

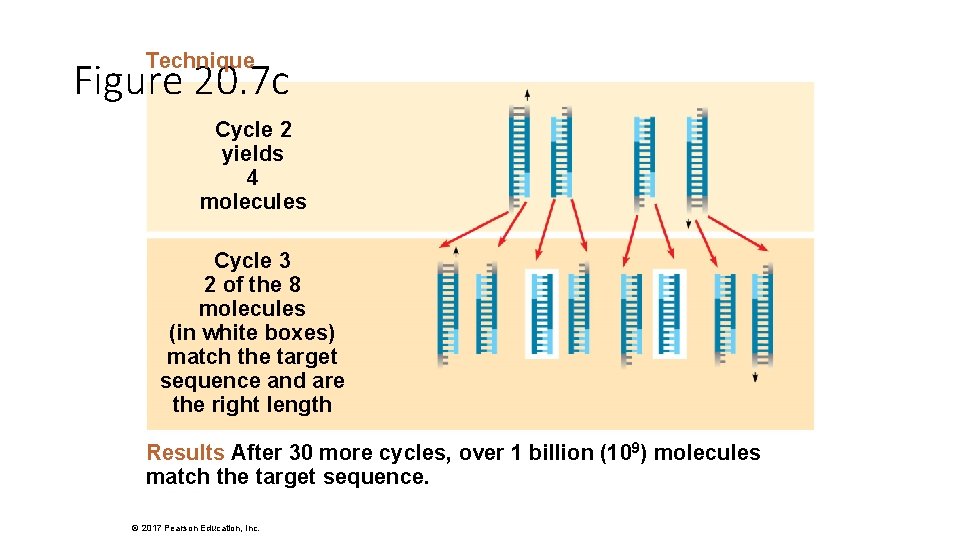
Technique Figure 20. 7 c Cycle 2 yields 4 molecules Cycle 3 2 of the 8 molecules (in white boxes) match the target sequence and are the right length Results After 30 more cycles, over 1 billion (109) molecules match the target sequence. © 2017 Pearson Education, Inc.

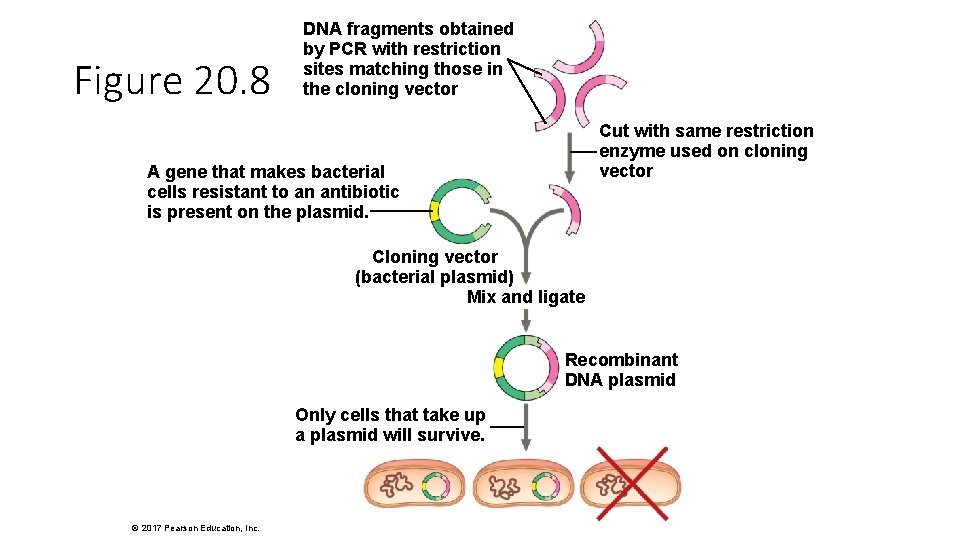
Figure 20. 8 DNA fragments obtained by PCR with restriction sites matching those in the cloning vector Cut with same restriction enzyme used on cloning vector A gene that makes bacterial cells resistant to an antibiotic is present on the plasmid. Cloning vector (bacterial plasmid) Mix and ligate Recombinant DNA plasmid Only cells that take up a plasmid will survive. © 2017 Pearson Education, Inc.

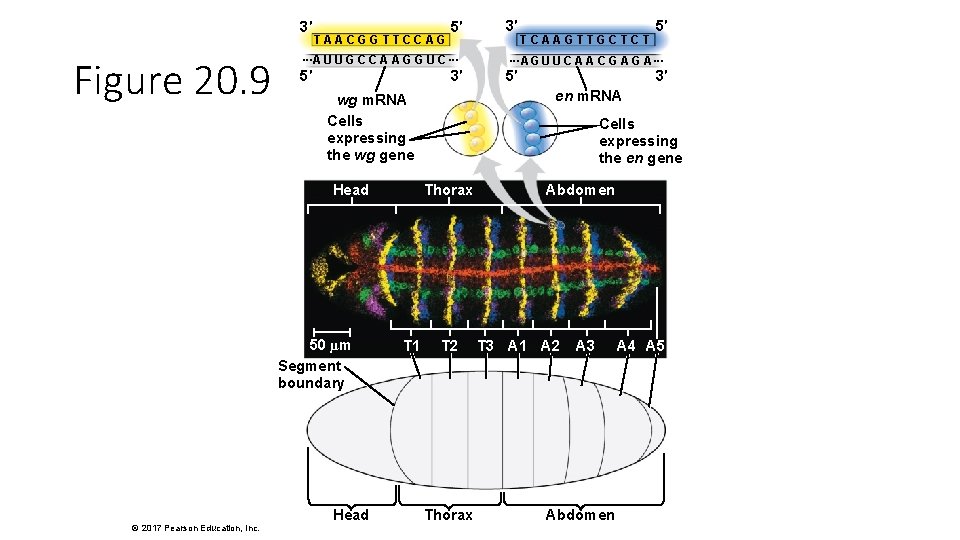
3′ Figure 20. 9 TAACGGTTCCAG ···A U U G C C A A G G U C ··· 5′ 3′ 50 µm Segment boundary Head TCAAGTTGCTCT 5′ ··· A G U U C A A C G A··· 5′ 3′ Cells expressing the en gene Thorax T 1 3′ en m. RNA wg m. RNA Cells expressing the wg gene Head © 2017 Pearson Education, Inc. 5′ T 2 Thorax Abdomen T 3 A 1 A 2 A 3 Abdomen A 4 A 5

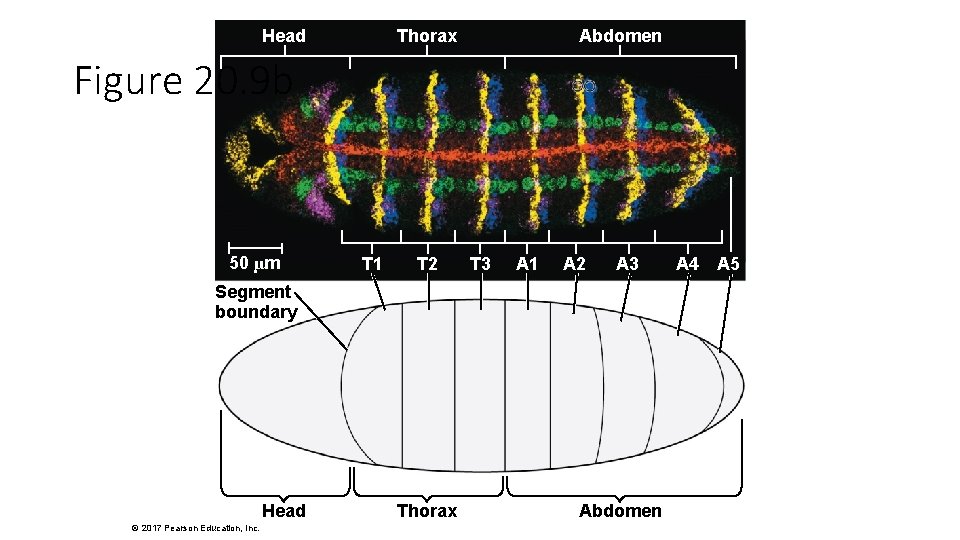
Head Thorax Abdomen Figure 20. 9 b 50 µm T 1 T 2 T 3 A 1 A 2 A 3 Segment boundary Head © 2017 Pearson Education, Inc. Thorax Abdomen A 4 A 5

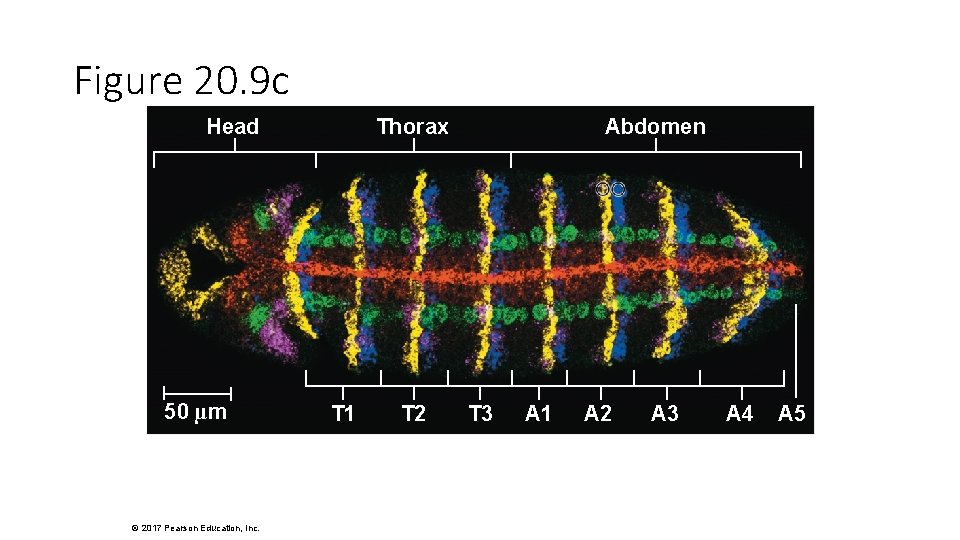
Figure 20. 9 c Head 50 µm © 2017 Pearson Education, Inc. Thorax T 1 T 2 Abdomen T 3 A 1 A 2 A 3 A 4 A 5

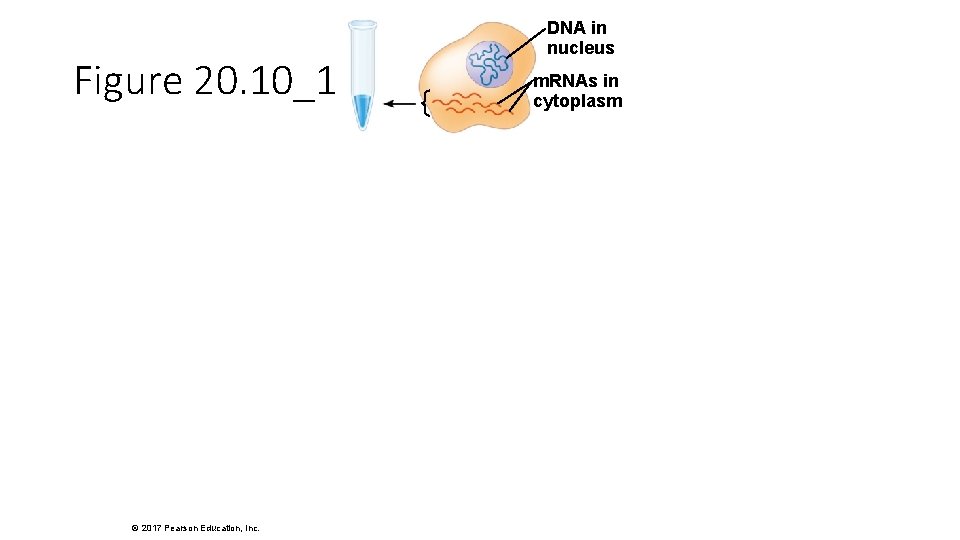
Figure 20. 10_1 © 2017 Pearson Education, Inc. DNA in nucleus m. RNAs in cytoplasm

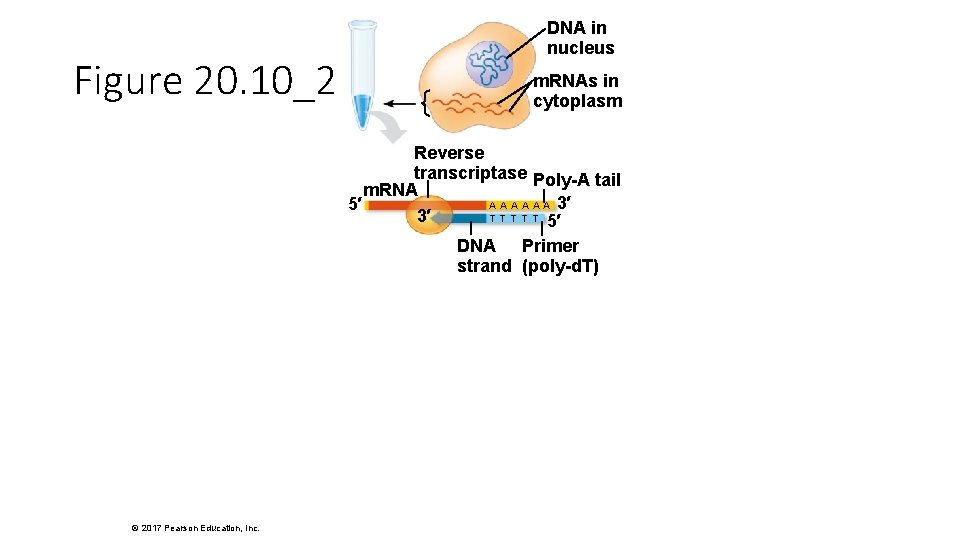
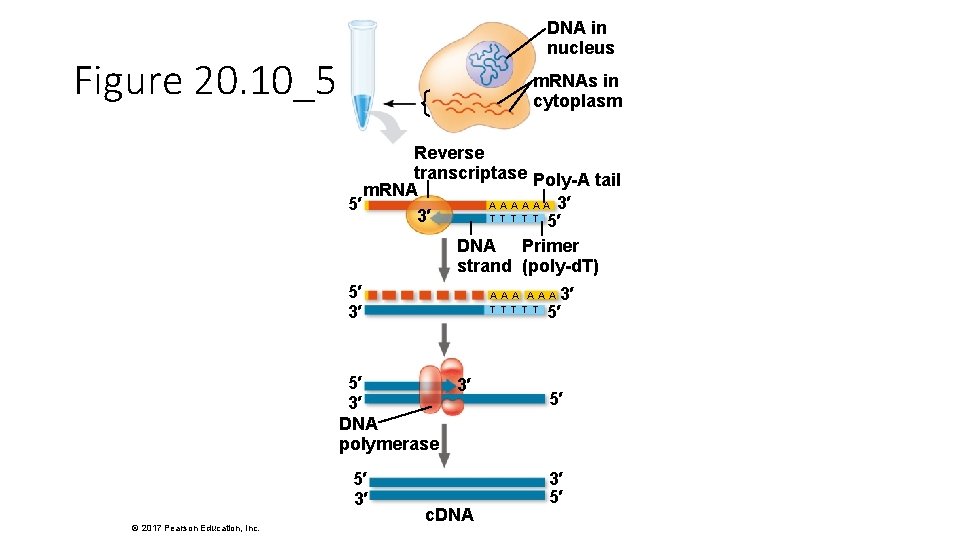
Figure 20. 10_2 DNA in nucleus m. RNAs in cytoplasm Reverse transcriptase Poly-A tail m. RNA A A A 3′ 5′ T T T 3′ 5′ DNA Primer strand (poly-d. T) © 2017 Pearson Education, Inc.

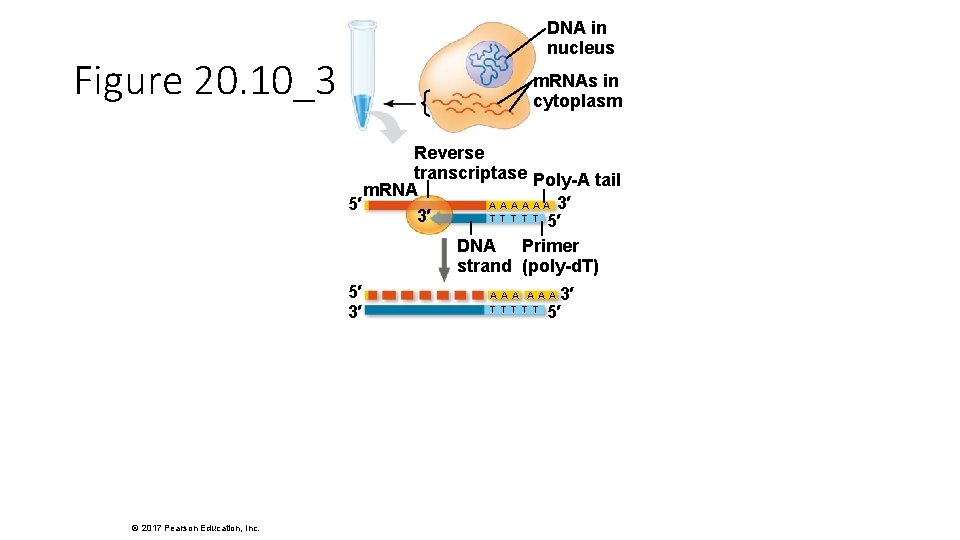
DNA in nucleus Figure 20. 10_3 m. RNAs in cytoplasm Reverse transcriptase Poly-A tail m. RNA A A A 3′ 5′ T T T 3′ 5′ DNA Primer strand (poly-d. T) 5′ 3′ © 2017 Pearson Education, Inc. 3′ 5′ AAA T T T

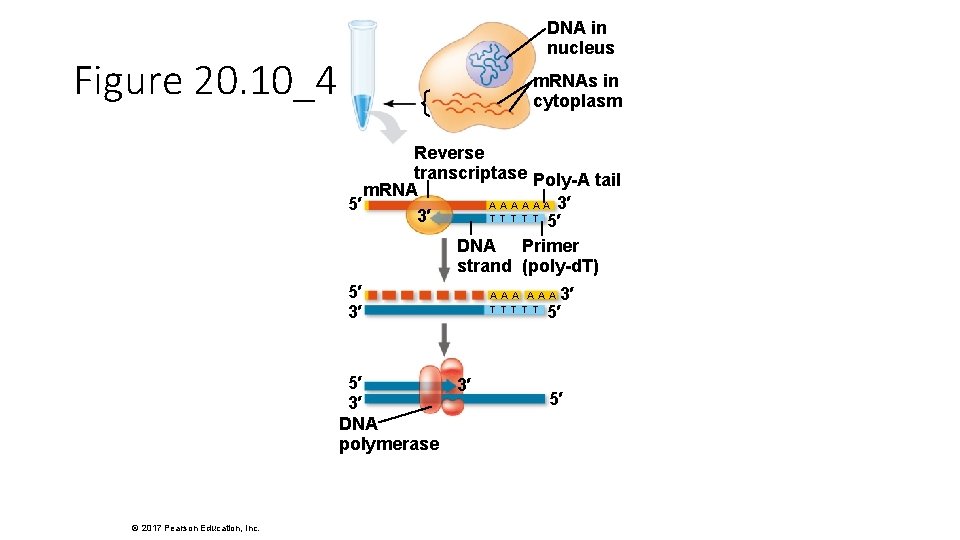
DNA in nucleus Figure 20. 10_4 m. RNAs in cytoplasm Reverse transcriptase Poly-A tail m. RNA A A A 3′ 5′ T T T 3′ 5′ DNA Primer strand (poly-d. T) 5′ 3′ DNA polymerase © 2017 Pearson Education, Inc. 3′ 5′ AAA T T T 3′ 5′

DNA in nucleus Figure 20. 10_5 m. RNAs in cytoplasm Reverse transcriptase Poly-A tail m. RNA A A A 3′ 5′ T T T 3′ 5′ DNA Primer strand (poly-d. T) 5′ 3′ 3′ 5′ AAA T T T 5′ 3′ DNA polymerase 5′ 3′ © 2017 Pearson Education, Inc. 3′ c. DNA 5′ 3′ 5′

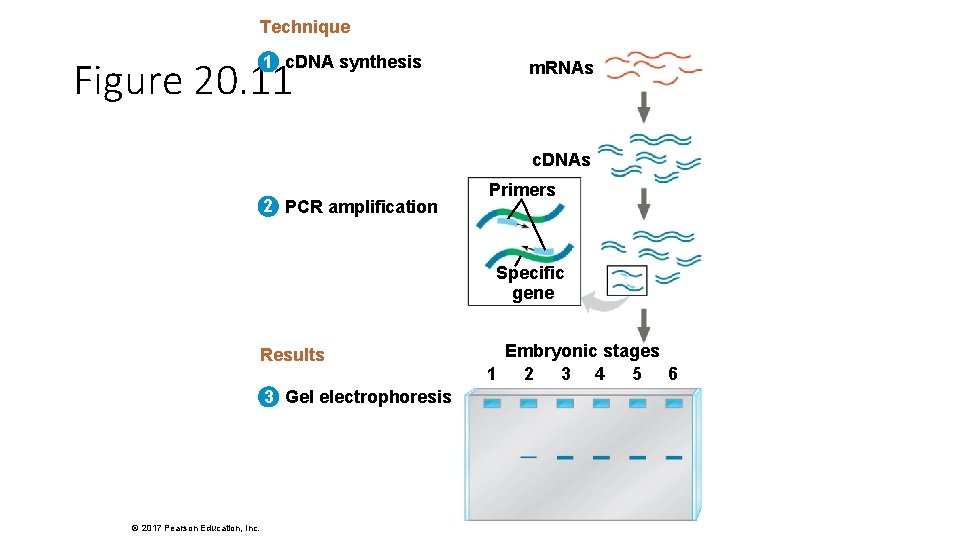
Technique 1 c. DNA synthesis Figure 20. 11 m. RNAs c. DNAs 2 PCR amplification Primers Specific gene Results 3 Gel electrophoresis © 2017 Pearson Education, Inc. Embryonic stages 1 2 3 4 5 6

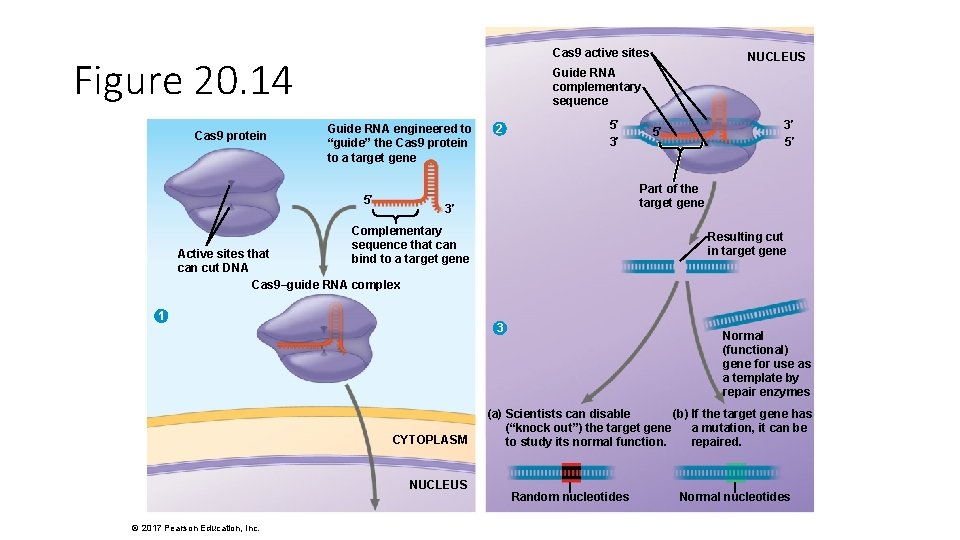
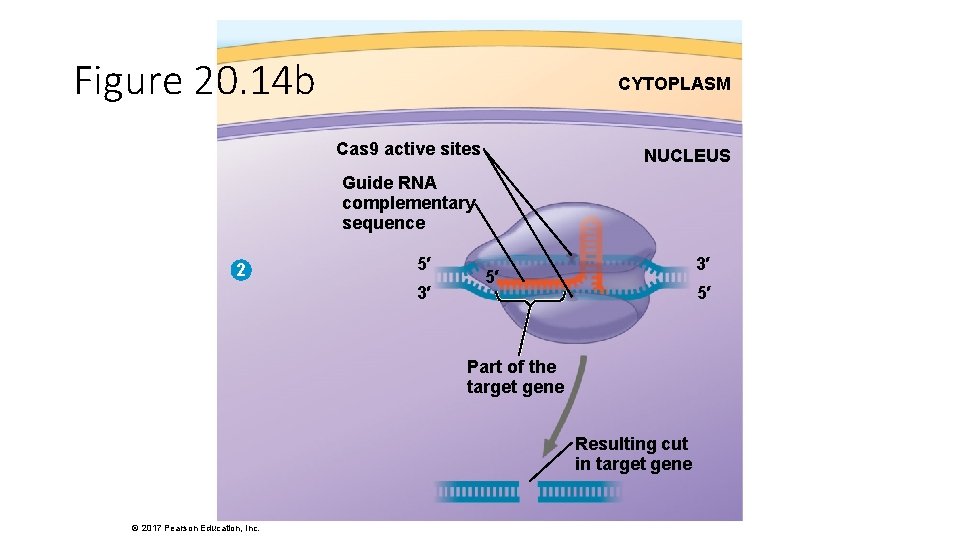
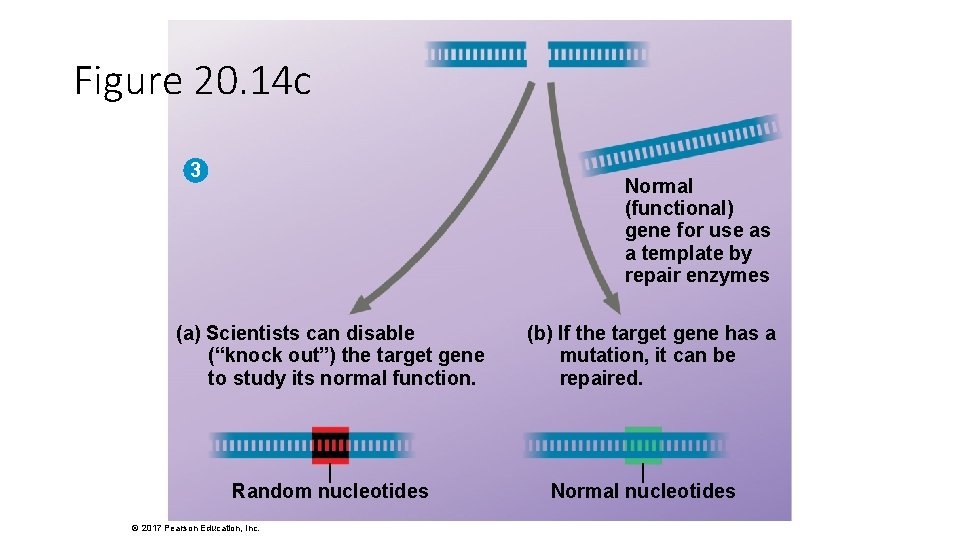
Cas 9 active sites Figure 20. 14 Cas 9 protein Guide RNA complementary sequence Guide RNA engineered to “guide” the Cas 9 protein to a target gene 5′ 2 5′ 3′ 3′ Resulting cut in target gene Active sites that can cut DNA Cas 9–guide RNA complex 1 3 CYTOPLASM NUCLEUS 3′ 5′ 5′ Part of the target gene Complementary sequence that can bind to a target gene © 2017 Pearson Education, Inc. NUCLEUS Normal (functional) gene for use as a template by repair enzymes (a) Scientists can disable (b) If the target gene has (“knock out”) the target gene a mutation, it can be to study its normal function. repaired. Random nucleotides Normal nucleotides

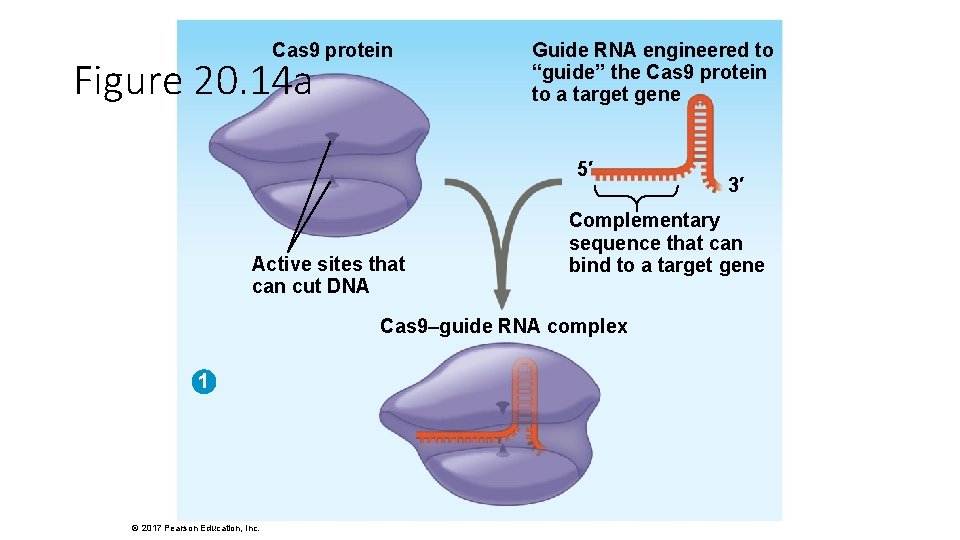
Cas 9 protein Figure 20. 14 a Guide RNA engineered to “guide” the Cas 9 protein to a target gene 5′ Active sites that can cut DNA Complementary sequence that can bind to a target gene Cas 9–guide RNA complex 1 © 2017 Pearson Education, Inc. 3′

Figure 20. 14 b CYTOPLASM Cas 9 active sites NUCLEUS Guide RNA complementary sequence 2 5′ 3′ 3′ 5′ 5′ Part of the target gene Resulting cut in target gene © 2017 Pearson Education, Inc.

Figure 20. 14 c 3 Normal (functional) gene for use as a template by repair enzymes (a) Scientists can disable (“knock out”) the target gene to study its normal function. Random nucleotides © 2017 Pearson Education, Inc. (b) If the target gene has a mutation, it can be repaired. Normal nucleotides

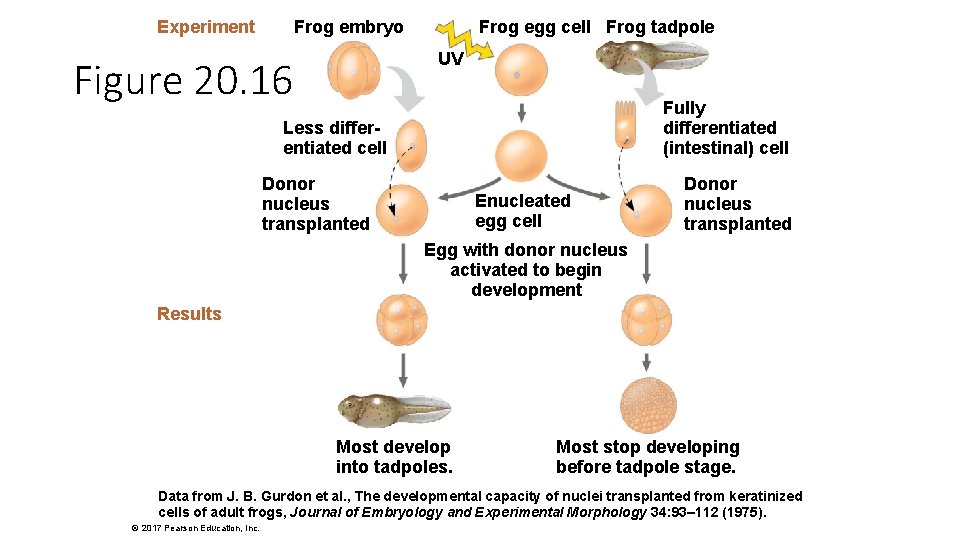
Experiment Frog embryo Frog egg cell Frog tadpole UV Figure 20. 16 Fully differentiated (intestinal) cell Less differentiated cell Donor nucleus transplanted Enucleated egg cell Donor nucleus transplanted Egg with donor nucleus activated to begin development Results Most develop into tadpoles. Most stop developing before tadpole stage. Data from J. B. Gurdon et al. , The developmental capacity of nuclei transplanted from keratinized cells of adult frogs, Journal of Embryology and Experimental Morphology 34: 93– 112 (1975). © 2017 Pearson Education, Inc.