Taxonomic Tree Tool A tool for managing and













- Slides: 30

Taxonomic Tree Tool - A tool for managing and comparing taxonomic trees Lin Contian 1, Qiao Huijie 1, Wang Jiangning 1, Ji Liqiang 1*, David Patterson 2 1 Institute of Zoology, CAS, 1 Beichen West Road, Chaoyang District, Beijing 100101, P. R. China 2 Biodiversity Informatics, Marine Biological Laboratory, Woods Hole, MA 02543, USA. * ji@ioz. ac. cn

Outline � Why Taxonomic Tree Tool (TTT)? � Taxonomic Tree Comparison � How Dose TTT Work?

Why TTT? � � � Taxonomic trees are important in the fields of taxonomy and biodiversity informatics. Taxonomic information is usually scattered in books, journals, specimen collections, and in the minds of people everywhere. Taxonomists need such a tool to gather data from different sources, organize the data, do research on it and share their own data or taxonomic views with others.

Why TTT? � � � Numerous taxonomic trees have been created due to different views, application of new technology like phylogenetics and different study geographic scopes. Taxonomists need a tool to explore the differences between their own taxonomic view and others’ for further study Biodiversity informatics initiatives would like to know what are the differences and then which one to pick up as backbone or how to integrate them

Why TTT? � � � Catalog of Life (COL) seeks to integrated distributed taxonomies into a single authoritative one. GBIF and GNA store trees in Checklist. Bank and Global Names Classification and List Repository (GNACLR) respectively. None of these projects has successfully been able to integrate the multiple taxonomies into a single flawless tree. Tree comparison will help identify consensus, point to areas of conflict, and promote progress towards large scale authoritative trees

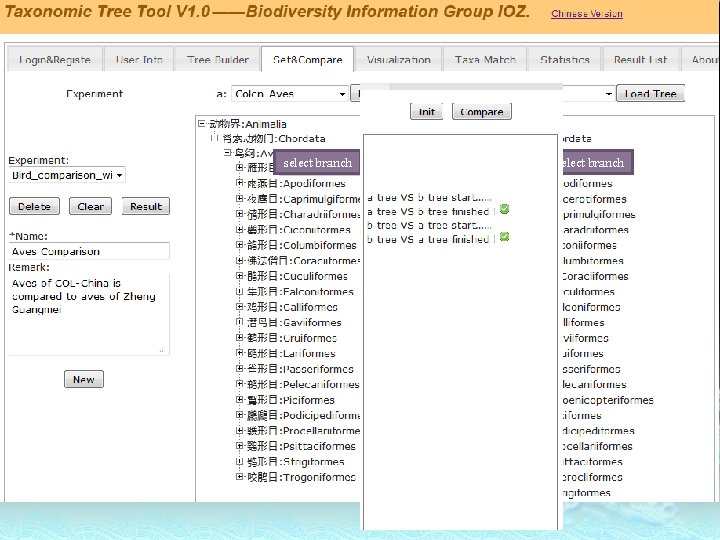
Taxonomic Tree Comparison

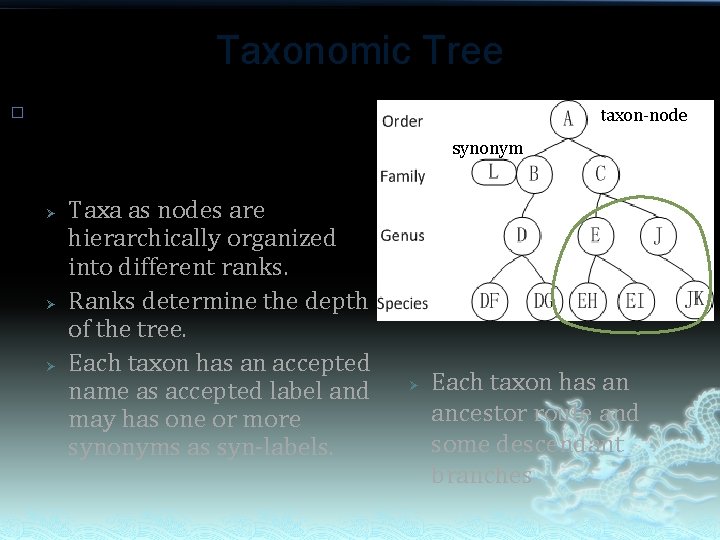
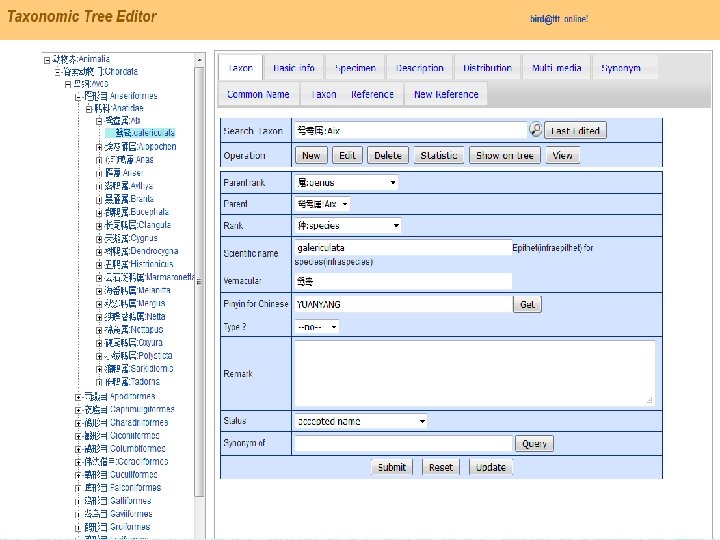
Taxonomic Tree � Technically, taxonomic tree is a special case of mathematical tree: Ø Ø Ø Taxa as nodes are hierarchically organized into different ranks. Ranks determine the depth of the tree. Each taxon has an accepted name as accepted label and may has one or more synonyms as syn-labels. taxon-node synonym Ø Each taxon has an ancestor route and some descendant branches

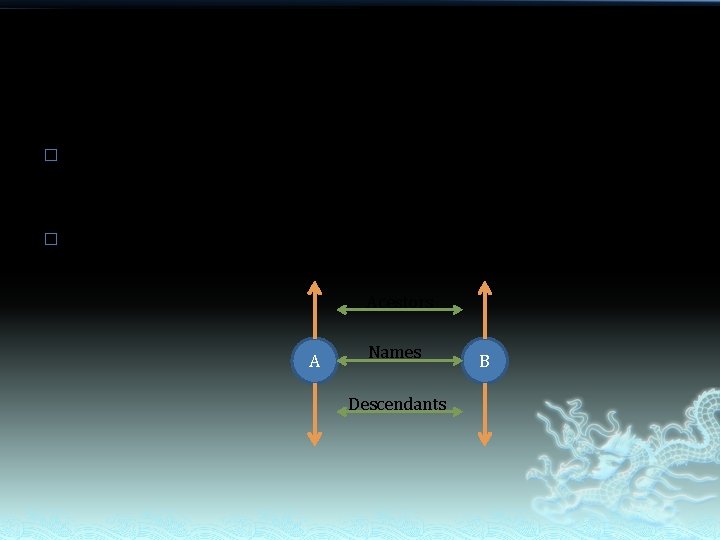
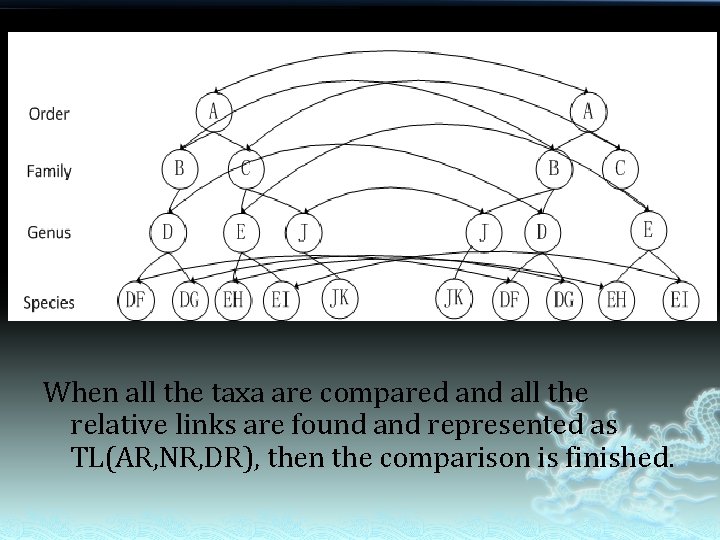
� � Since taxon is the unit of taxonomic tree, we should compare taxa when we compare tree. What of the taxa should be compared? Acestors A Names Descendants B

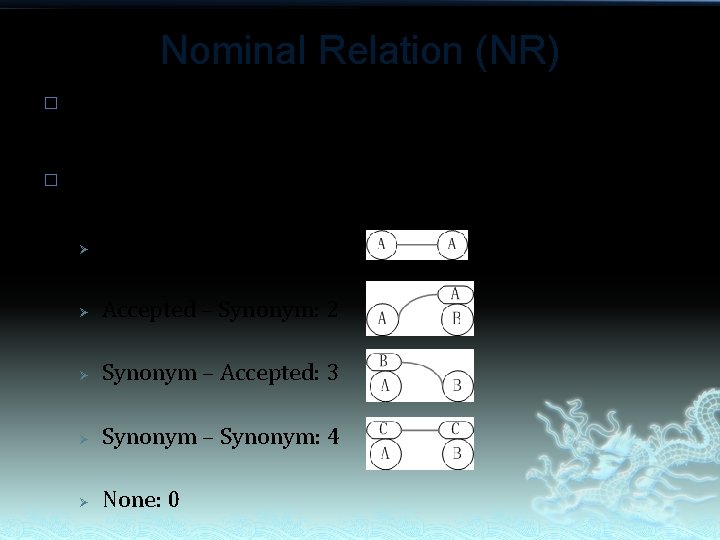
Nominal Relation (NR) � � Scientific names are keys to a taxon though they cannot uniquely represent a taxon. Nominal Relations between two taxa should be: Ø Accepted – Accepted: 1 Ø Accepted – Synonym: 2 Ø Synonym – Accepted: 3 Ø Synonym – Synonym: 4 Ø None: 0

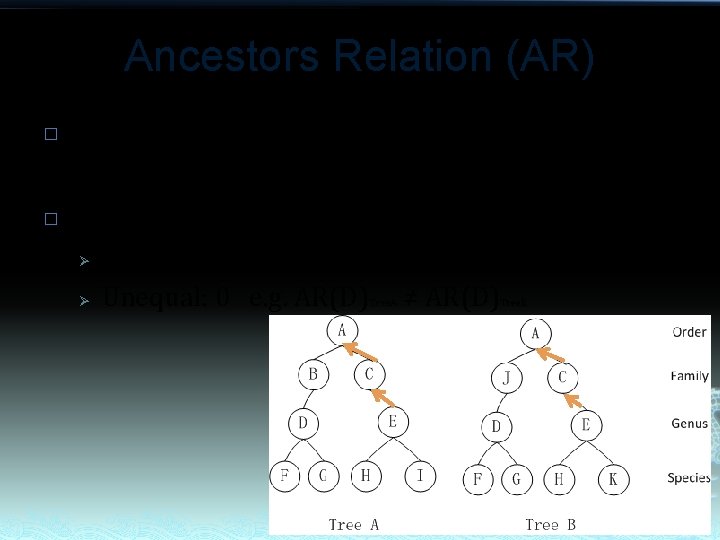
Ancestors Relation (AR) � � AR reflects difference of taxonomic position from top down between two taxa. Value of AR should be: Ø Ø Equal: 1 e. g. AR(E)Tree. A= AR(E) Unequal: 0 e. g. AR(D) ≠ AR(D) Tree. B Tree. A Tree. B

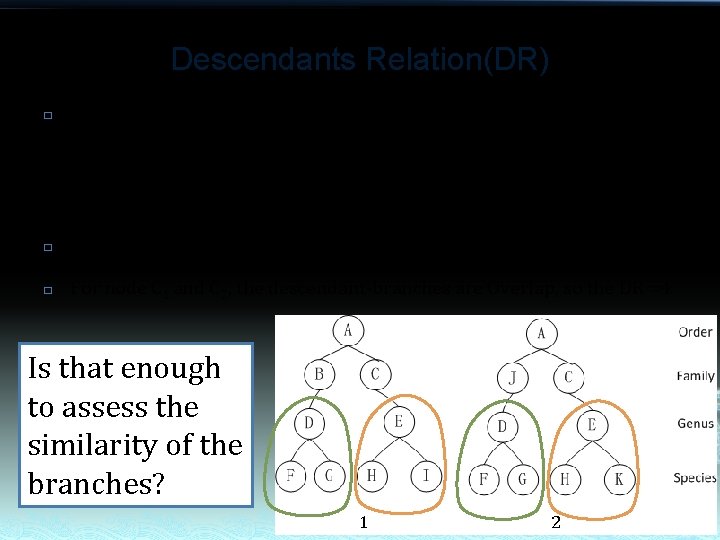
Descendants Relation(DR) � There are five status of DR: Congruent, DR=1 Included, DR=2 Include, DR=3 Overlap, DR=4 Exclude, DR=0 � � For node B 1 and J 2, the descendant-branches are congruent, so the DR =1 For node C 1 and C 2, the descendant-branches are Overlap, so the DR =4 Is that enough to assess the similarity of the branches? 1 2

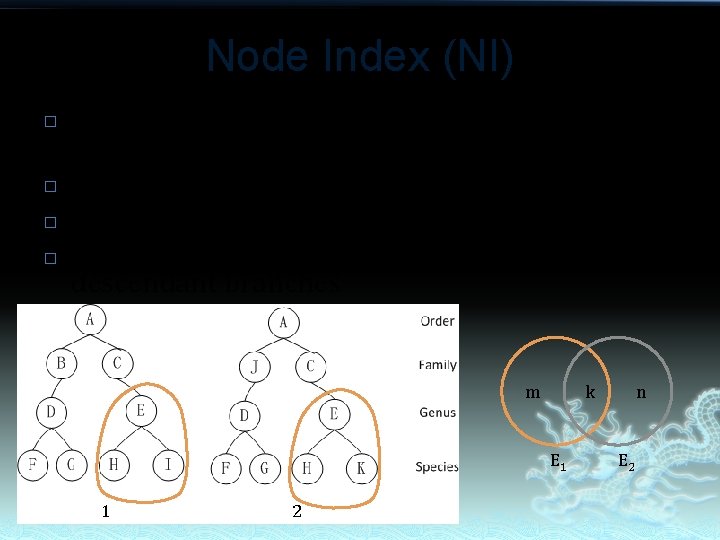
Node Index (NI) � � Node Index is calculated as the ratio of the common nodes. For node E 1, NI(E 1)=k/m=2/3 For node E 2, NI(E 2)=k/n=2/3 NIs reflect the similarity of nodes in descendant branches m k E 1 1 2 n E 2

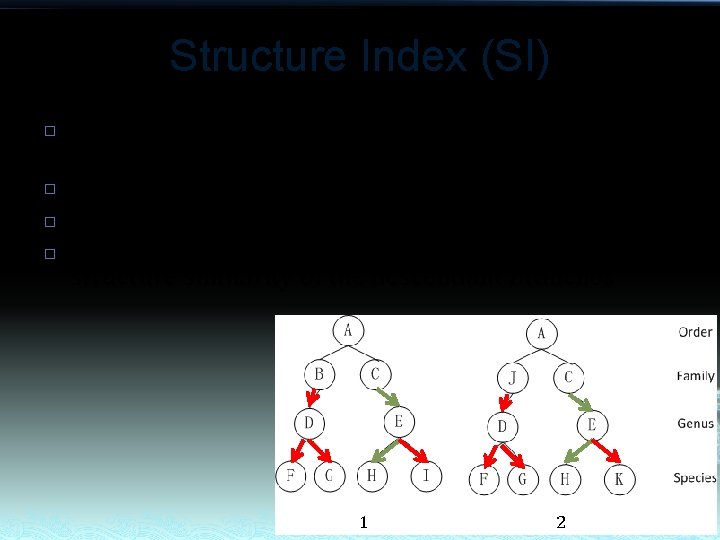
Structure Index (SI) � � Structure Index is calculated as ratio of common routes. SI(A 1 )=1/4 SI(A 2 )=1/4 Structure Index is intended for assessing the structure similarity of the descendant branches 1 2

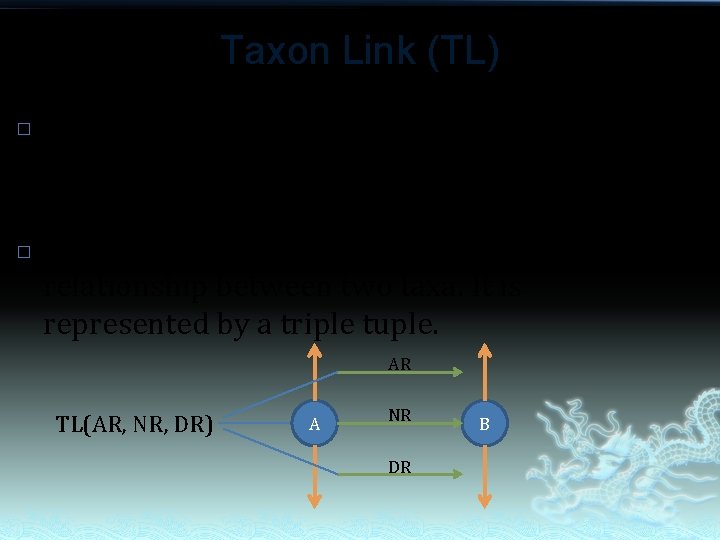
Taxon Link (TL) � � What we can do with Nominal Relation (NR), Ancestors Relation(AR) , Descendants Relation(DR)? Yes, Taxon Link! Taxon Link is intended for describe the relationship between two taxa. It is represented by a triple tuple. AR TL(AR, NR, DR) A NR DR B

When all the taxa are compared and all the relative links are found and represented as TL(AR, NR, DR), then the comparison is finished.

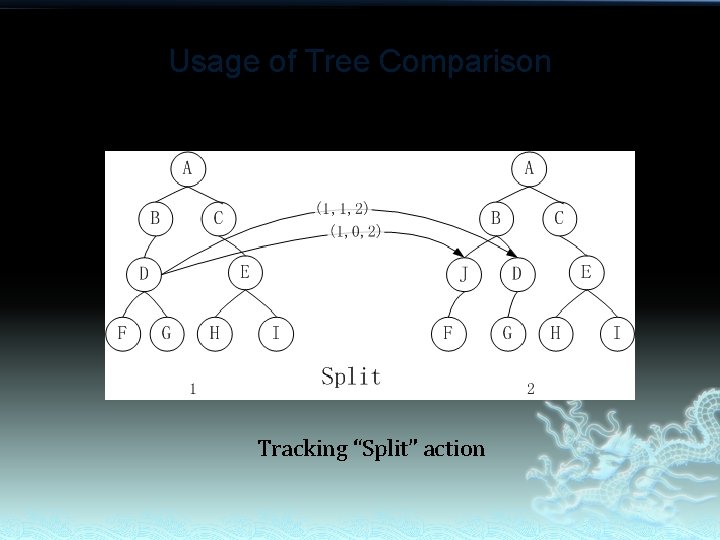
Usage of Tree Comparison Tracking “Split” action

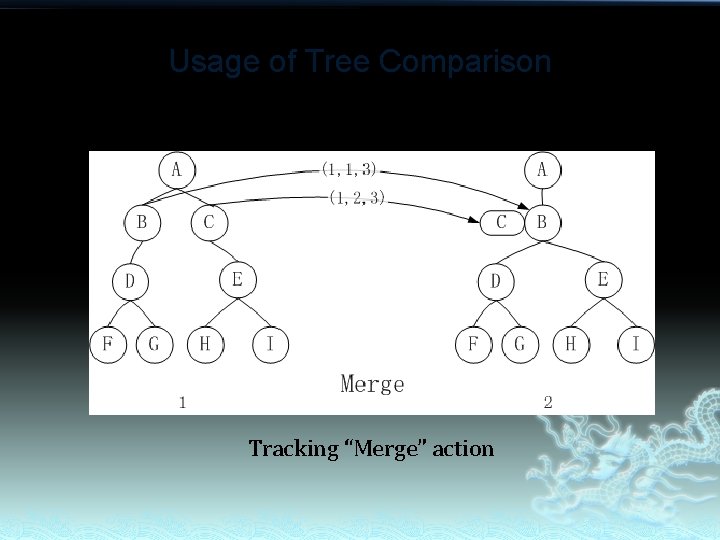
Usage of Tree Comparison Tracking “Merge” action

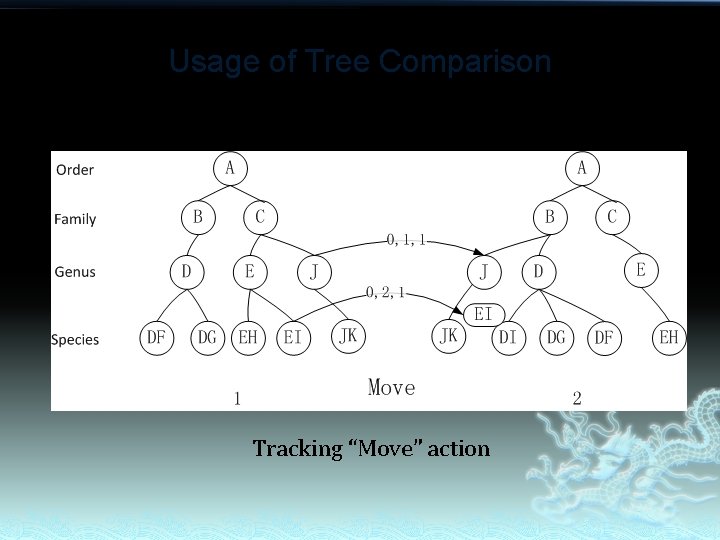
Usage of Tree Comparison Tracking “Move” action

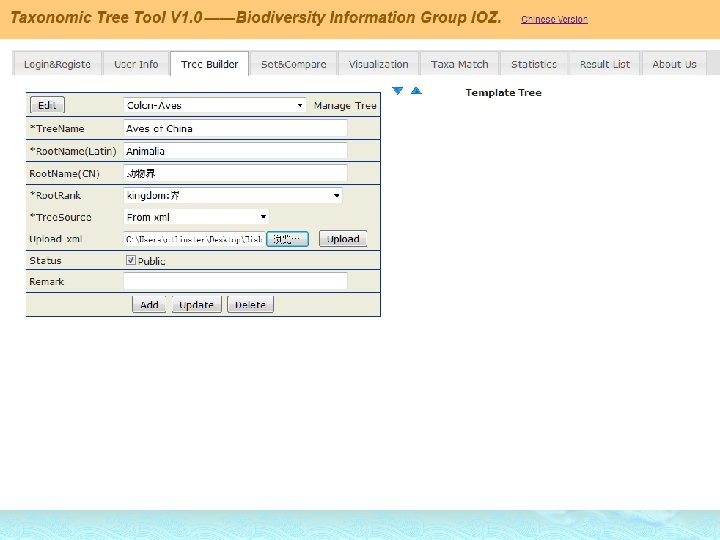
How Dose TTT Work?

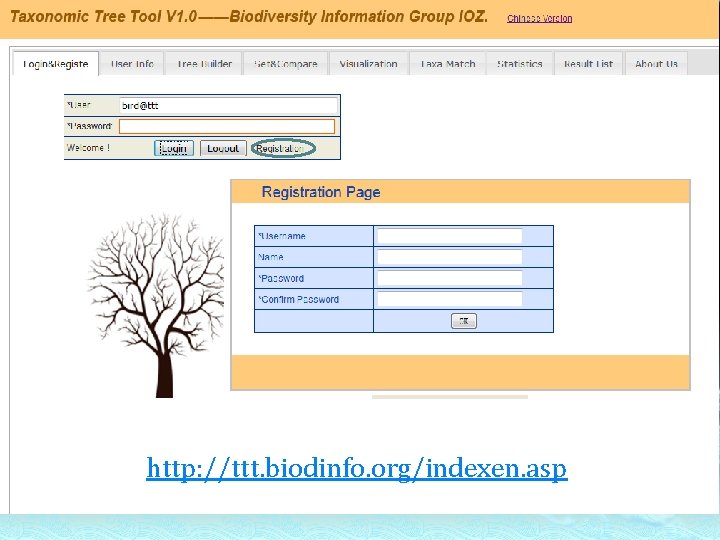
http: //ttt. biodinfo. org/indexen. asp







select branch

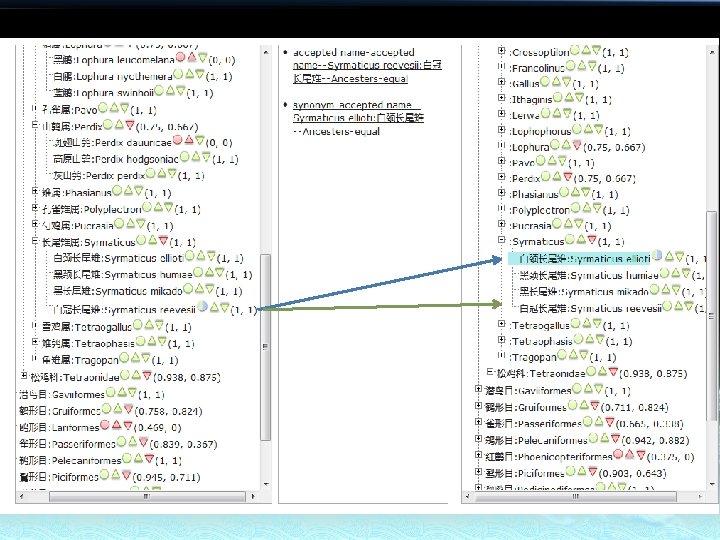
Tree A Taxon Link tuple is represented by three different shapes. Tree B Nominal Relation Ancestor Relation Descendants Relation


Thank you for your attention! � http: //ttt. biodinfo. org � Feed back to linct@ioz. ac. cn �