Tandem Mass SpectrometryBased Approaches For the Characterization of

Tandem Mass Spectrometry-Based Approaches For the Characterization of Glycans and Glycopeptides Lance Wells, Jae-Min Lim, Dan Sherling, Bryan Woosley, Dawei Lin, Ron Orlando, Enas El-Karim, Chin-Fin Teo, Olga Stuchlik, Michael Tiemeyer& Carl Bergman Complex Carbohydrate Research Center, Department of Biochemistry & Molecular Biology University of Georgia
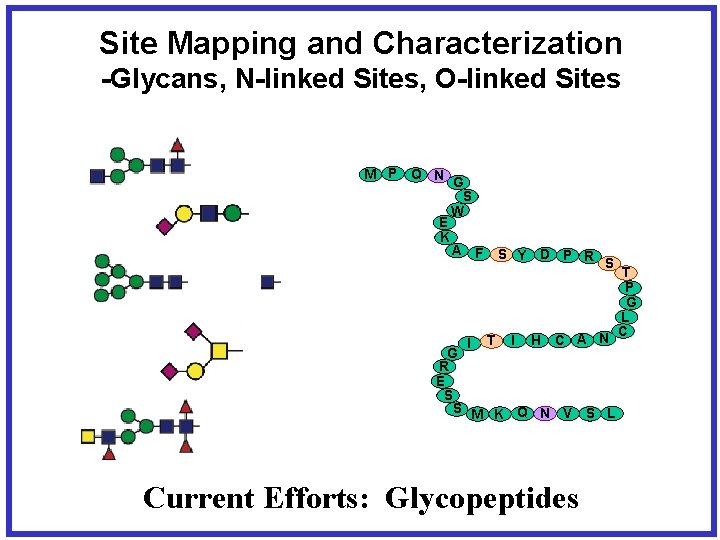
Site Mapping and Characterization -Glycans, N-linked Sites, O-linked Sites M P Q N G S W E K A F I S Y D P R S T I H C A N G R E S S M K Q N V S L Current Efforts: Glycopeptides T P G L C
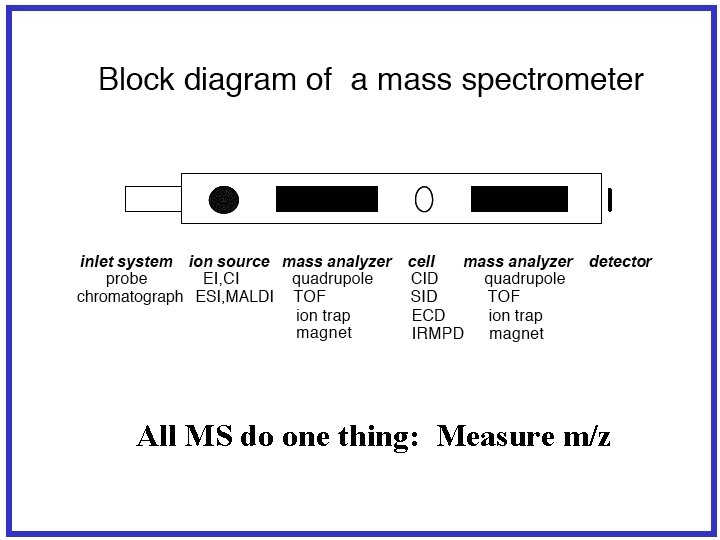
All MS do one thing: Measure m/z
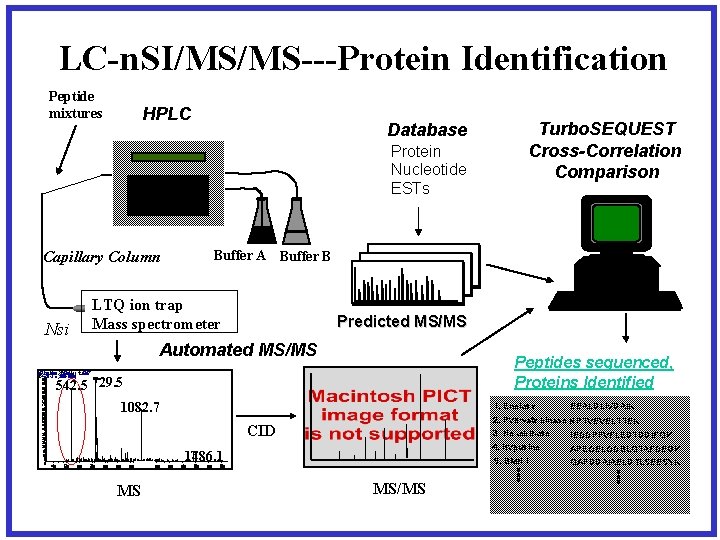
LC-n. SI/MS/MS---Protein Identification Peptide mixtures HPLC Database Protein Nucleotide ESTs Buffer A Buffer B Capillary Column Nsi LTQ ion trap Mass spectrometer Predicted MS/MS Automated MS/MS 2 M ar 3 #u 2 ll m 183 R T 0 : 3 T 5 : + c N 1 SM I F s [ 35. 050. 9 -14 A 750 V. 0: 1 N 0] L: 1. 56 E 7 729. 5 100 95 542. 8 90 85 80 75 70 65 60 55 50 45 40 35 30 25 20 15 10 5 0 400 500 600 700 800 Peptides sequenced, Proteins Identified 542. 5 729. 5 1082. 7 1. Enolase 1082. 7 4. Hypusine 1486. 1 1000 1100 MS 1200 1300 1400 1500 1600 EEALDLIVDAIK 2. Pyruvate kinase NPTVEVELTTEK 3. Hexokinase IEDDPFVFLEDTDDIFQK CID 900 Turbo. SEQUEST Cross-Correlation Comparison 5. BMH 1 1700 MS/MS APEGELGDSLQTAFDEGK QAFDDAIAELDTLSEESYK
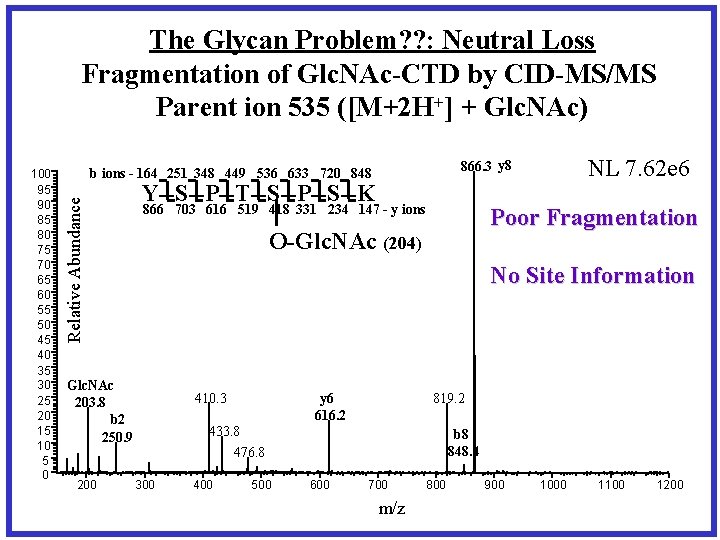
The Glycan Problem? ? : Neutral Loss Fragmentation of Glc. NAc-CTD by CID-MS/MS Parent ion 535 ([M+2 H+] + Glc. NAc) Y--S--P--T--S--P--S--K 866 703 616 519 418 331 234 147 - y ions Poor Fragmentation O-Glc. NAc (204) No Site Information Glc. NAc 203. 8 b 2 250. 9 200 NL 7. 62 e 6 866. 3 y 8 b ions - 164 251 348 449 536 633 720 848 Relative Abundance 100 95 90 85 80 75 70 65 60 55 50 45 40 35 30 25 20 15 10 5 0 410. 3 y 6 616. 2 819. 2 433. 8 476. 8 300 400 500 b 8 848. 4 600 700 m/z 800 900 1000 1100 1200
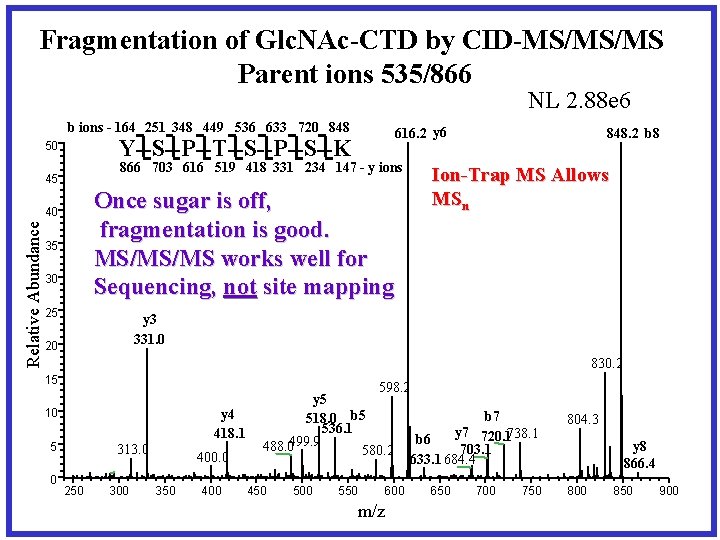
Fragmentation of Glc. NAc-CTD by CID-MS/MS/MS Parent ions 535/866 NL 2. 88 e 6 b ions - 164 251 348 449 536 633 720 848 866 703 616 519 418 331 234 147 - y ions 45 Once sugar is off, fragmentation is good. MS/MS/MS works well for Sequencing, not site mapping 40 Relative Abundance 616. 2 y 6 Y--S--P--T--S--P--S--K 50 35 30 25 848. 2 b 8 Ion-Trap MS Allows MSn y 3 331. 0 20 830. 2 15 y 4 418. 1 10 5 0 313. 0 250 300 400. 0 350 400 598. 2 y 5 b 7 518. 0 b 5 536. 1 y 7 720. 1738. 1 b 6 488. 0499. 9 703. 1 580. 2 633. 1 684. 4 450 500 550 600 m/z 650 700 750 804. 3 y 8 866. 4 800 850 900
What We Really Need? • A method that allows for mapping of sites • A method that allows for enrichment of modified peptides • A method that can be used to do quantitative mass spectrometry (isotope labeling of PTM)
BEMAD Methodology for Sites of O-Glycosylation
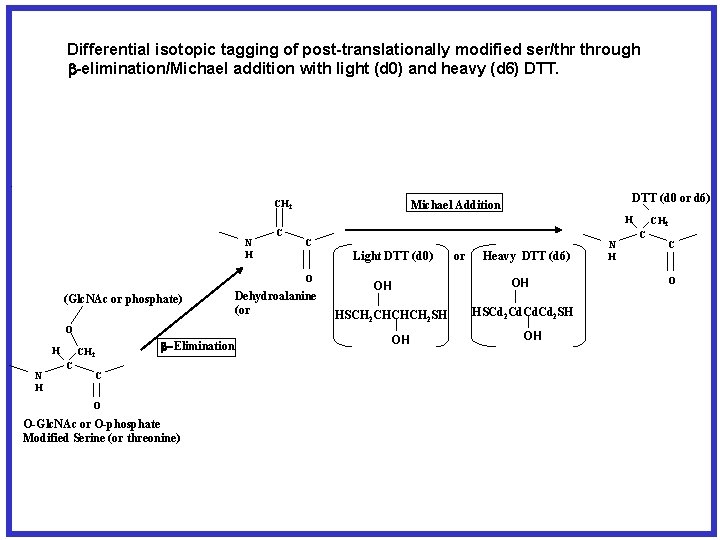
Differential isotopic tagging of post-translationally modified ser/thr through b-elimination/Michael addition with light (d 0) and heavy (d 6) DTT. O S H CH 2 C N H C NH 2 b-Elimination C CH 2 H O Alkylated Cysteine N H C C Light DTT (d 0) O (Glc. NAc or phosphate) O H N H CH 2 C DTT (d 0 or d 6) Michael Addition b-Elimination C O O-Glc. NAc or O-phosphate Modified Serine (or threonine) Dehydroalanine (or or Heavy DTT (d 6) OH OH HSCH 2 CHCHCH 2 SH OH HSCd 2 Cd. Cd 2 SH OH N H CH 2 C C O
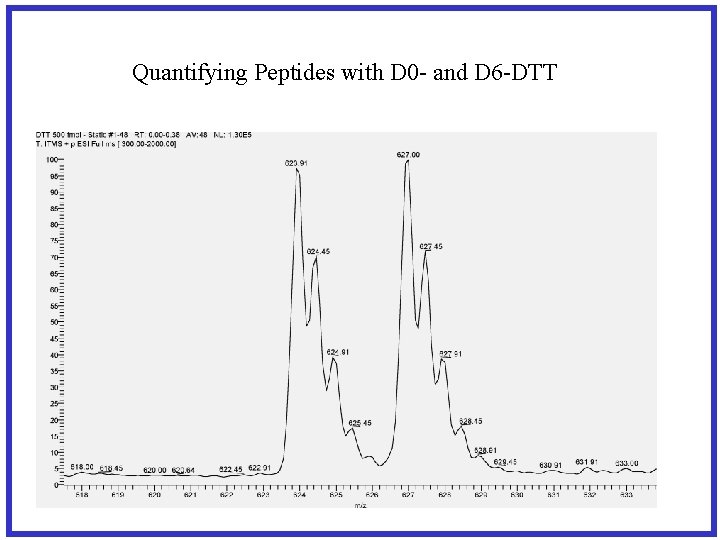
Quantifying Peptides with D 0 - and D 6 -DTT
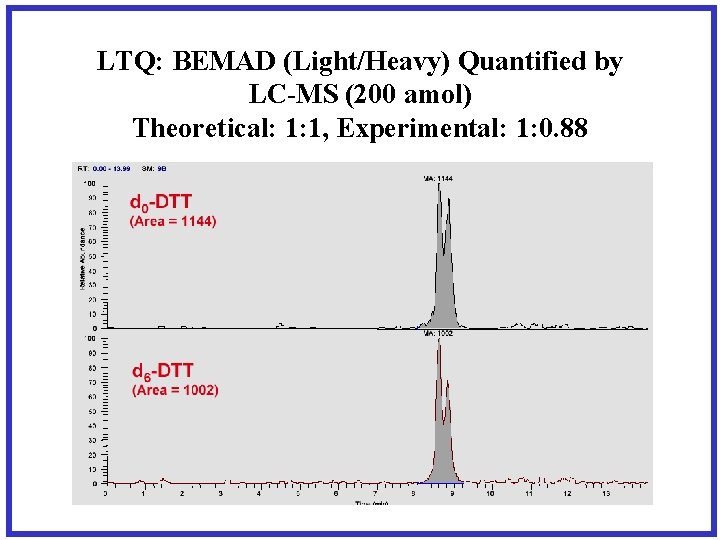
LTQ: BEMAD (Light/Heavy) Quantified by LC-MS (200 amol) Theoretical: 1: 1, Experimental: 1: 0. 88
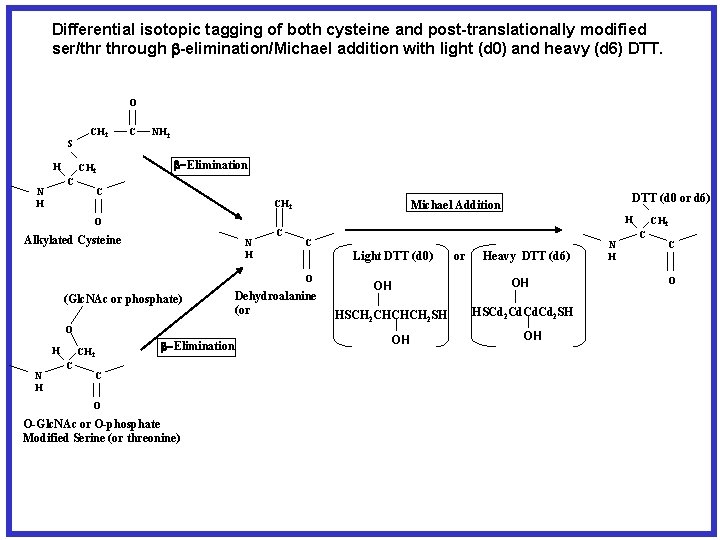
Differential isotopic tagging of both cysteine and post-translationally modified ser/thr through b-elimination/Michael addition with light (d 0) and heavy (d 6) DTT. O S H CH 2 C N H C NH 2 b-Elimination C CH 2 H O Alkylated Cysteine N H C C Light DTT (d 0) O (Glc. NAc or phosphate) O H N H CH 2 C DTT (d 0 or d 6) Michael Addition b-Elimination C O O-Glc. NAc or O-phosphate Modified Serine (or threonine) Dehydroalanine (or or Heavy DTT (d 6) OH OH HSCH 2 CHCHCH 2 SH OH HSCd 2 Cd. Cd 2 SH OH N H CH 2 C C O
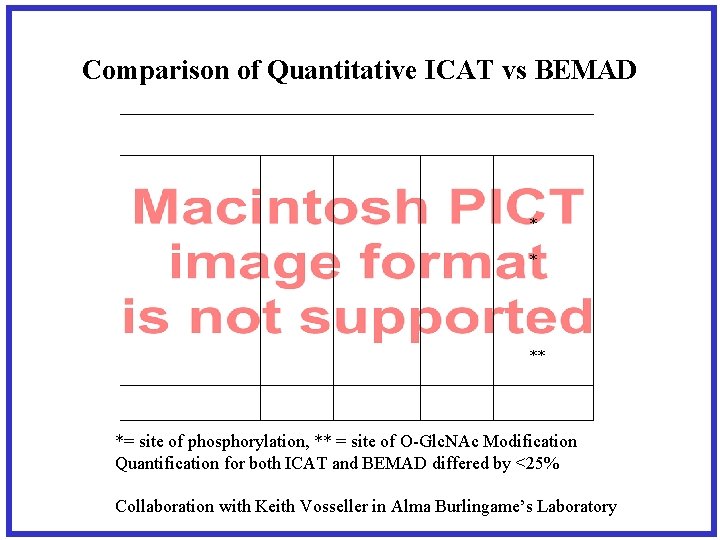
Comparison of Quantitative ICAT vs BEMAD * * ** *= site of phosphorylation, ** = site of O-Glc. NAc Modification Quantification for both ICAT and BEMAD differed by <25% Collaboration with Keith Vosseller in Alma Burlingame’s Laboratory
BEMAD and Neutral Loss Approaches for Other O-Glycans • O-Man (not extended) on fungal proteins • Complex O-Glycosylation
Neutral Loss and BEMAD Mapping of O-Man Sites on PGC
BEMAD for Mapping Complex O-Glycosylation
ECD Fragmentation Site-Mapping on an LTQ-FT
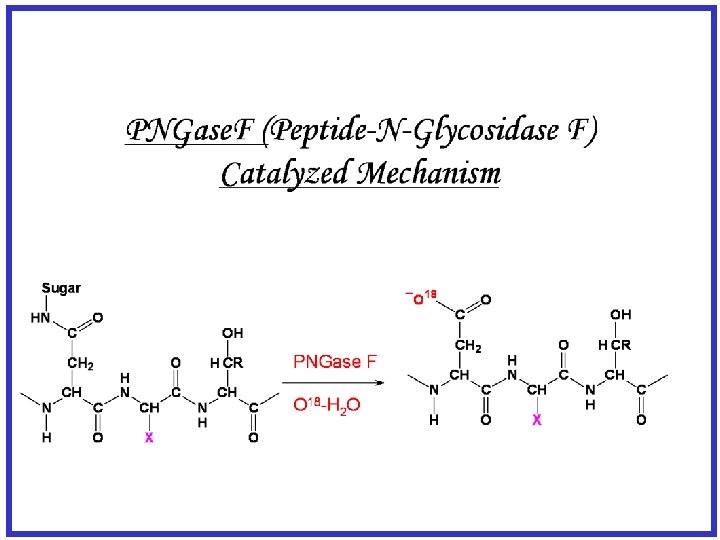

N-linked glycosylation on Secreted Adipocyte Proteome Site mapping using PNGase. F + 18 O Water Sulfated glycoprotein 1 precursor (SGP-1) gi|3914939 (K)TVVTEAGNLLKDN#ATQEEILHYLEK (K)FSELIVNN#ATEELLVK (K)LVLYLEHNLEKN#STKEEILAALEK Follistatin-related protein 1 precursor gi|2498391 (K)GSN#YSEILDK Haptoglobin gi|8850219 (K)VVLHPN#HSVVDIGLIK (K)NLFLN#HSETASAK (K)CVVHYEN#STVPEKK lipoprotein lipase gi|12832783 (R)TPEDTAEDTCHLIPGLADSVSNCHFN#HSSK vimentin gi|2078001 (R)QVQSLTCEVDALKGTN#ESLER Adipsin gi|673431 (K)LSQN#ASLGPHVRPLPLQYEDK Decorin gi|6681143 Hemopexin gi|1881768 (R)ISDTN#ITAIPQGLPTSLTEVHLDGNK (R)SWSTVGN#CTAALR Cyclophilin C-associated protein MAMA/Cy. CAP gi|6755144 RIKEN c. DNA 9330129 D 05 gene gi|30520293 (K)GLN#LTEDTYKPR (R)ALGYEN#ATQALGR (K)MELKN#QSRLQEPAAR Cathepsin L gi|929719 PPBG/ Cathepsin A gi|12860234 (R)AEFAVAN#DTGFVDIPQQEK (R)LDPPCTN#TTAPSNYLNNPYVR Relative Quantification Possible by 16 O and 18 O Water
Characterizing Plant Protein PGIP • N-linked glycoprotein • 7 Putative Sites of Glycosylation • Collaboration with Carl Bergmann’s and Mike Tiemeyer’s group at the CCRC

LC-MS/MS Analysis of O-18 labeled PGIP
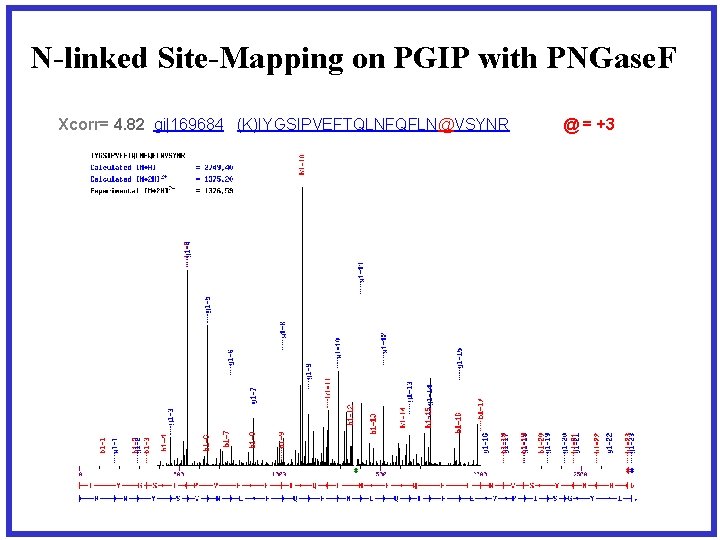
N-linked Site-Mapping on PGIP with PNGase. F Xcorr= 4. 82 gi|169684 (K)IYGSIPVEFTQLNFQFLN@VSYNR @ = +3
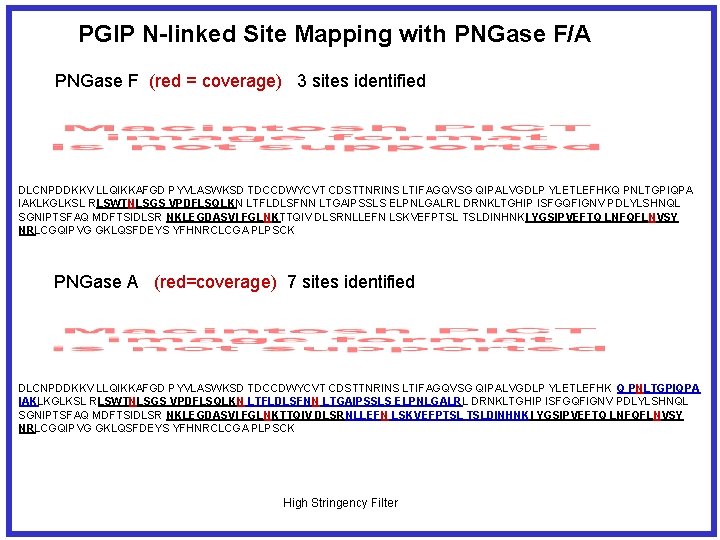
PGIP N-linked Site Mapping with PNGase F/A PNGase F (red = coverage) 3 sites identified DLCNPDDKKV LLQIKKAFGD PYVLASWKSD TDCCDWYCVT CDSTTNRINS LTIFAGQVSG QIPALVGDLP YLETLEFHKQ PNLTGPIQPA IAKLKGLKSL RLSWTNLSGS VPDFLSQLKN LTFLDLSFNN LTGAIPSSLS ELPNLGALRL DRNKLTGHIP ISFGQFIGNV PDLYLSHNQL SGNIPTSFAQ MDFTSIDLSR NKLEGDASVI FGLNKTTQIV DLSRNLLEFN LSKVEFPTSL TSLDINHNKI YGSIPVEFTQ LNFQFLNVSY NRLCGQIPVG GKLQSFDEYS YFHNRCLCGA PLPSCK PNGase A (red=coverage) 7 sites identified DLCNPDDKKV LLQIKKAFGD PYVLASWKSD TDCCDWYCVT CDSTTNRINS LTIFAGQVSG QIPALVGDLP YLETLEFHK Q PNLTGPIQPA IAKLKGLKSL RLSWTNLSGS VPDFLSQLKN LTFLDLSFNN LTGAIPSSLS ELPNLGALRL DRNKLTGHIP ISFGQFIGNV PDLYLSHNQL SGNIPTSFAQ MDFTSIDLSR NKLEGDASVI FGLNKTTQIV DLSRNLLEFN LSKVEFPTSL TSLDINHNKI YGSIPVEFTQ LNFQFLNVSY NRLCGQIPVG GKLQSFDEYS YFHNRCLCGA PLPSCK High Stringency Filter
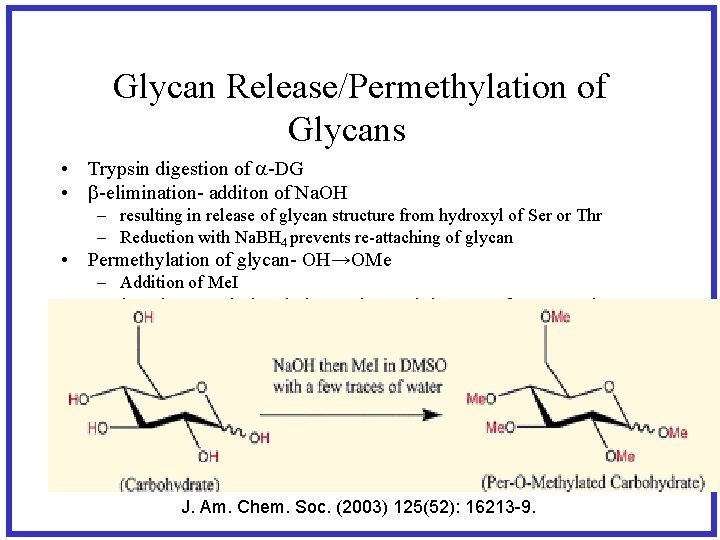
Glycan Release/Permethylation of Glycans • Trypsin digestion of a-DG • b-elimination- additon of Na. OH – resulting in release of glycan structure from hydroxyl of Ser or Thr – Reduction with Na. BH 4 prevents re-attaching of glycan • Permethylation of glycan- OH→OMe – Addition of Me. I • Analyzed permethylated glycans by applying MSn fragmentation as needed to completely determine the structure J. Am. Chem. Soc. (2003) 125(52): 16213 -9.

LTQ PNGF 1331. 7 MALDI Permethylated PNGA 1505. 0
PNGase. F

1331 PNGase. F M 3 N 2 X Glc. NAc-OMe -Xyl, -H 2 O Glc. NAc-OMe -Glc. NAc- - Me. OH

1505 PNGase. A
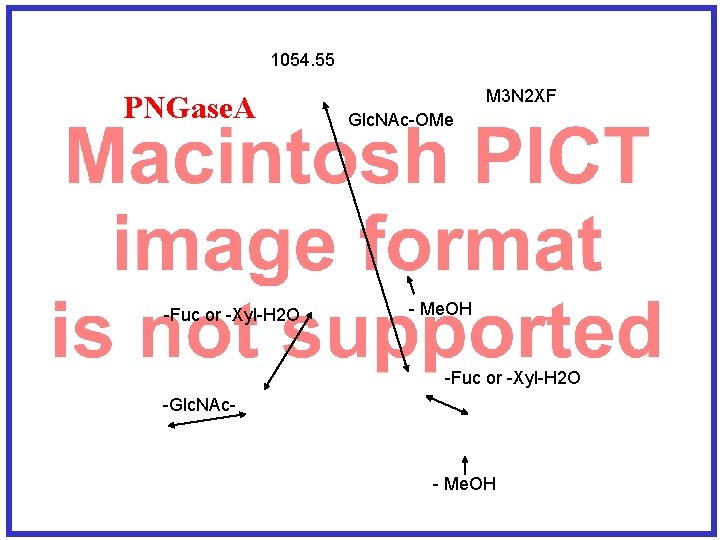
1054. 55 PNGase. A -Fuc or -Xyl-H 2 O M 3 N 2 XF Glc. NAc-OMe - Me. OH -Fuc or -Xyl-H 2 O -Glc. NAc- - Me. OH
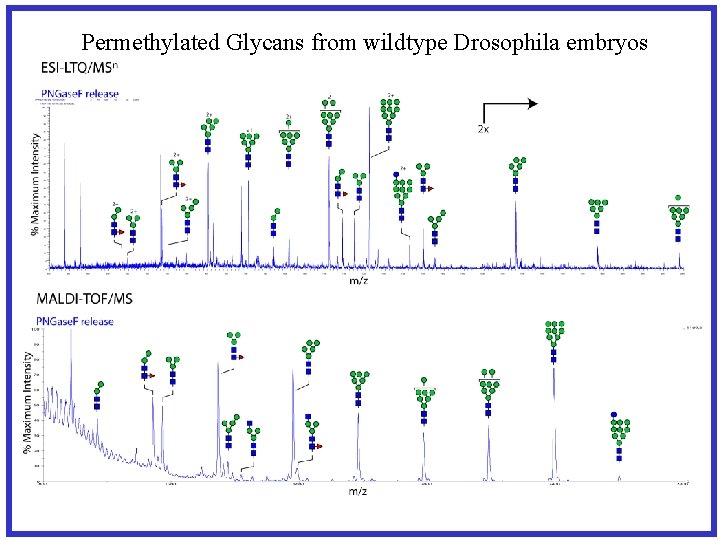
Permethylated Glycans from wildtype Drosophila embryos
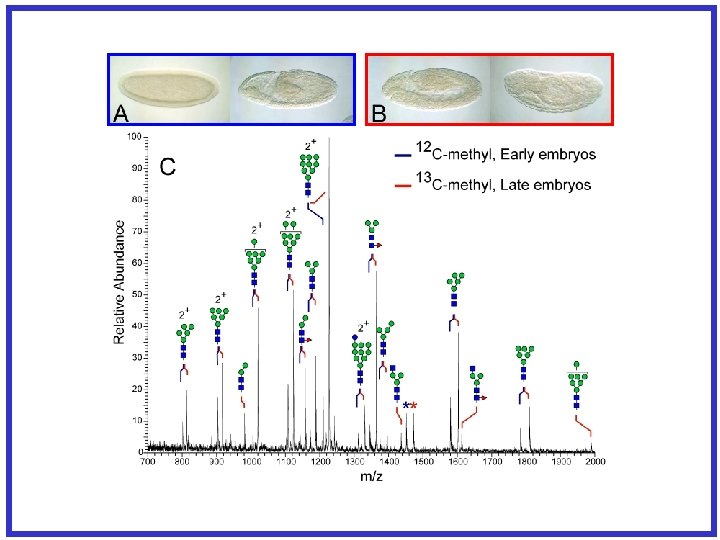
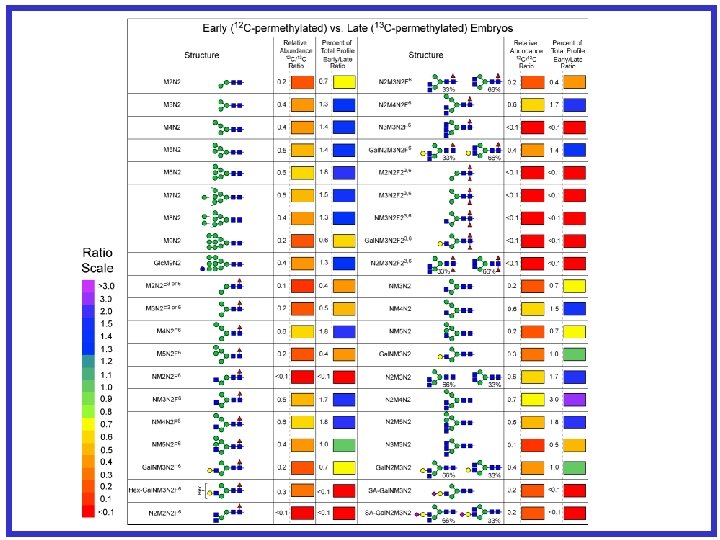
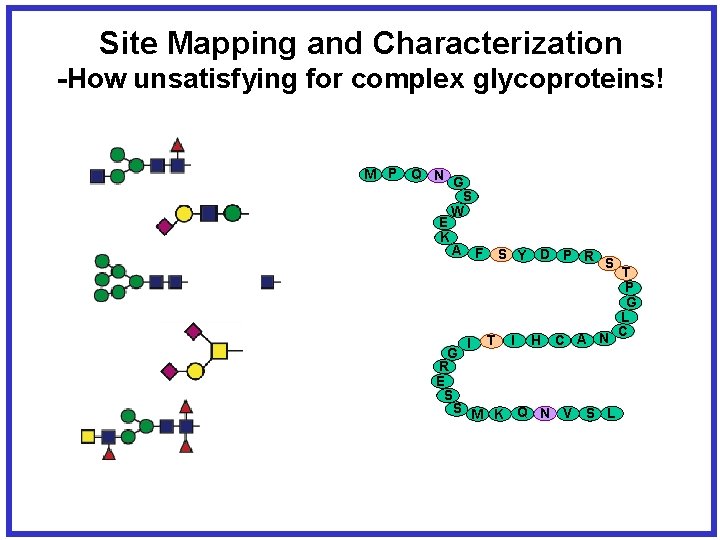
Site Mapping and Characterization -How unsatisfying for complex glycoproteins! M P Q N G S W E K A F I S Y D P R S T I H C A N G R E S S M K Q N V S L T P G L C
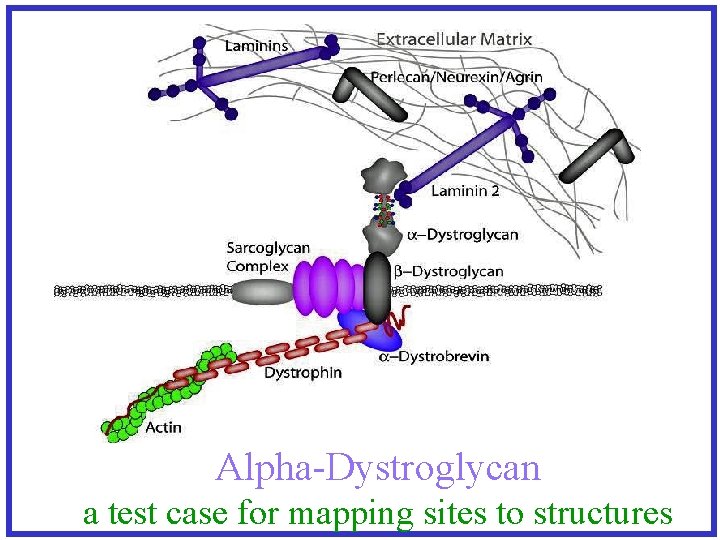
Alpha-Dystroglycan a test case for mapping sites to structures
Fragmentation of O-glycans -Hex-Hex. NAc + Na -SA -Hex
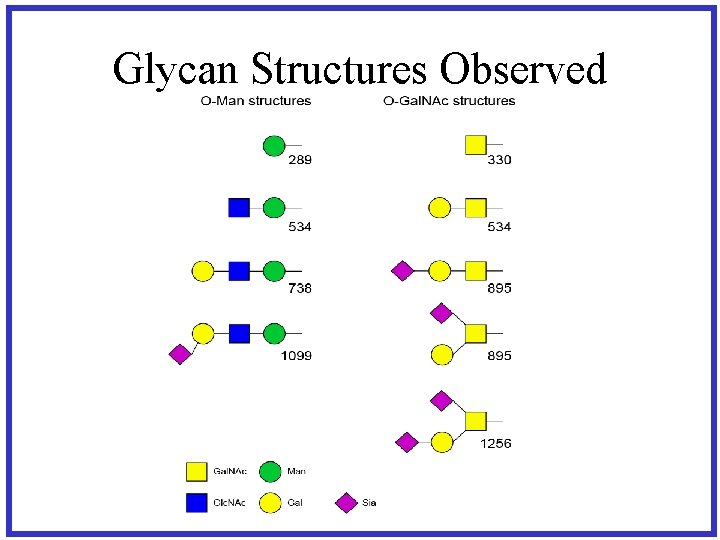
Glycan Structures Observed
3 MS • • – Pseudo Neutral Loss Overall survey scan 8 most intense peaks were selected Fragmentation was induced upon these peaks If result from fragmentation produced neutral loss, additional fragmentation was induced when the loss of a neutral from table was observed • Fragmentation was repeated resulting in MS 3
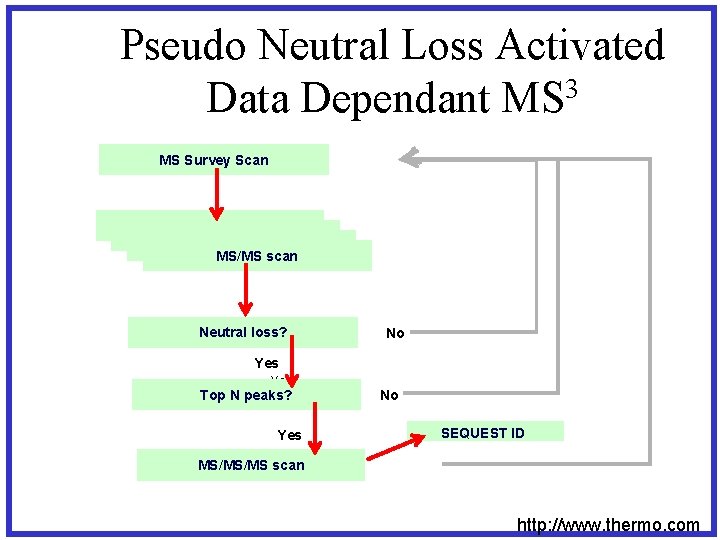
Pseudo Neutral Loss Activated Data Dependant MS 3 MS Survey Scan MS survey scan MS/MS scan Neutral loss? No Yes Top N peaks? Yes No SEQUEST ID MS/MS/MS scan http: //www. thermo. com

Use of Pseudo Neutral Loss Method Full Scan at 5. 81 minutes

MS/MS of 740. 25 Neutral loss of SA
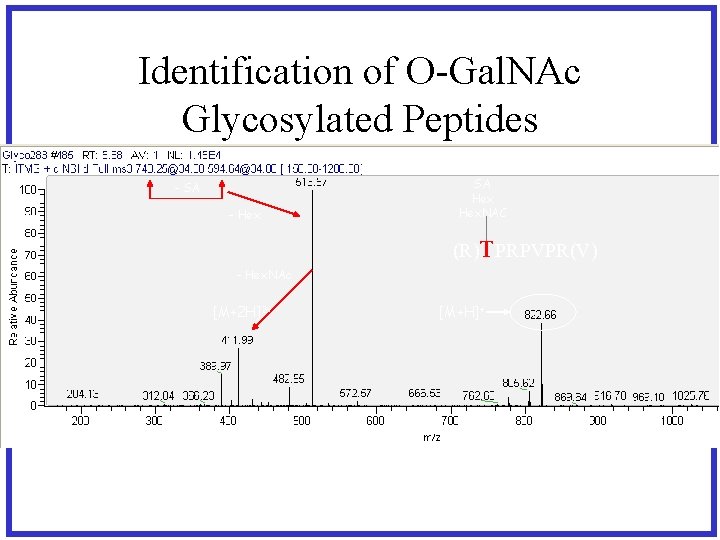
Identification of O-Gal. NAc Glycosylated Peptides - SA - Hex SA Hex. NAC (R)TPRPVPR(V) - Hex. NAc [M+2 H]2+ [M+H]+
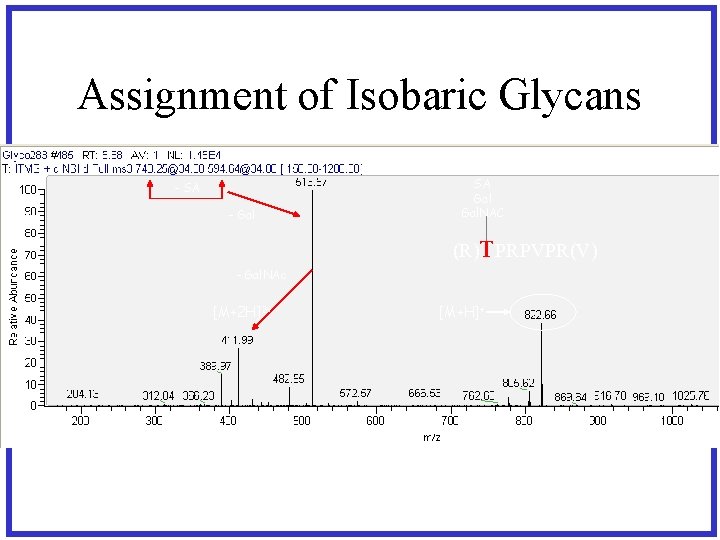
Assignment of Isobaric Glycans - SA - Gal SA Gal. NAC (R)TPRPVPR(V) - Gal. NAc [M+2 H]2+ [M+H]+
Full Scan at 52. 38 minutes
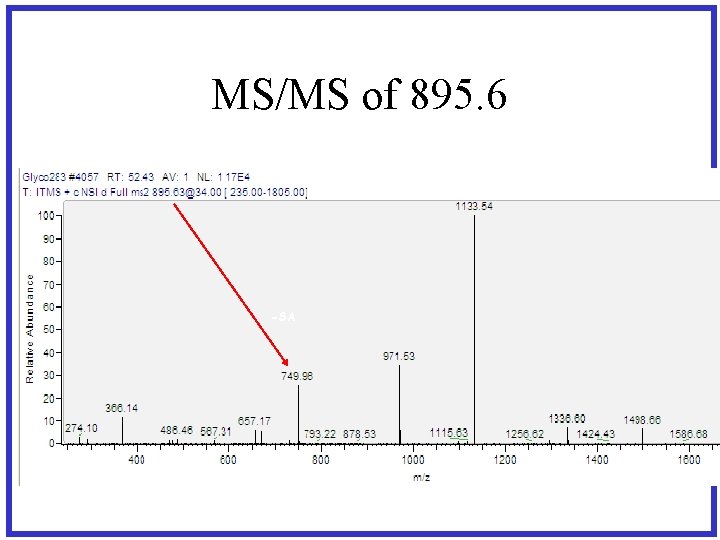
MS/MS of 895. 6 -SA
![Identification of O-Man Glycosylated Peptides SA Hex. NAc Hex -Hex (R)LETASPPTR(I) [M+H]+ [M+2 H]2+ Identification of O-Man Glycosylated Peptides SA Hex. NAc Hex -Hex (R)LETASPPTR(I) [M+H]+ [M+2 H]2+](http://slidetodoc.com/presentation_image_h/f45c877c930ee9ec744d14defd4a98c6/image-46.jpg)
Identification of O-Man Glycosylated Peptides SA Hex. NAc Hex -Hex (R)LETASPPTR(I) [M+H]+ [M+2 H]2+ -Hex. NAc
![Assignment of O-Man Isobaric Glycans SA Gal. NAc Man -Gal (R)LETASPPTR(I) [M+H]+ [M+2 H]2+ Assignment of O-Man Isobaric Glycans SA Gal. NAc Man -Gal (R)LETASPPTR(I) [M+H]+ [M+2 H]2+](http://slidetodoc.com/presentation_image_h/f45c877c930ee9ec744d14defd4a98c6/image-47.jpg)
Assignment of O-Man Isobaric Glycans SA Gal. NAc Man -Gal (R)LETASPPTR(I) [M+H]+ [M+2 H]2+ -Man -Gal. NAc
How can we isolate the exact site of glycosylation? • For peptide with only one Ser or Thr residue within sequence site of glycosylation is obvious • For peptides containing more that one Ser or Thr residue assignment is difficult… – Identify glycosylated peptide based upon addition of parent mass and combined neutral losses – Observe glycosylation of b & y ions – BEMAD-b-elimination and Michael addition of DTT
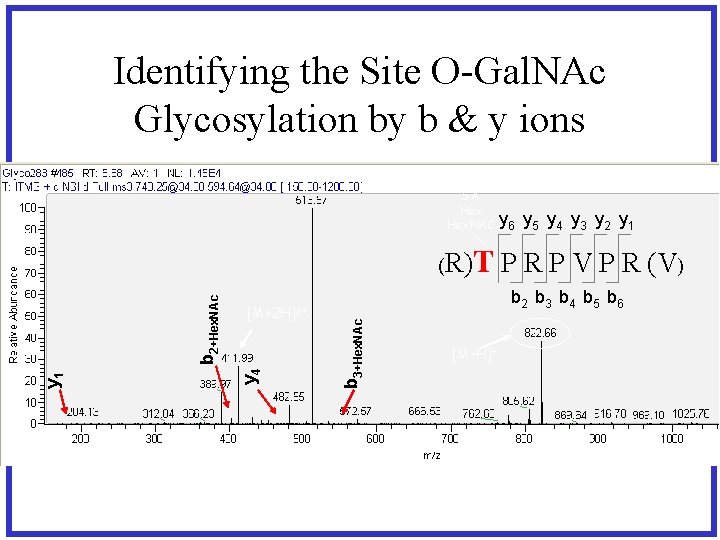
Identifying the Site O-Gal. NAc Glycosylation by b & y ions SA Hex. NAC T P R P V P R (V) b 2 b 3 b 4 b 5 b 6 b 3+Hex. NAc [M+2 H]2+ y 4 y 1 b 2+Hex. NAc (R) y 6 y 5 y 4 y 3 y 2 y 1 [M+H]+
![Identifying the Site O-Man Glycosylation by b & y ions -Hex. NAc [M+H]+ b Identifying the Site O-Man Glycosylation by b & y ions -Hex. NAc [M+H]+ b](http://slidetodoc.com/presentation_image_h/f45c877c930ee9ec744d14defd4a98c6/image-50.jpg)
Identifying the Site O-Man Glycosylation by b & y ions -Hex. NAc [M+H]+ b 6+Hex [M+2 H]2+ (R)LETASPPTR(I) b 5+Hex -Hex SA Hex. NAc Hex
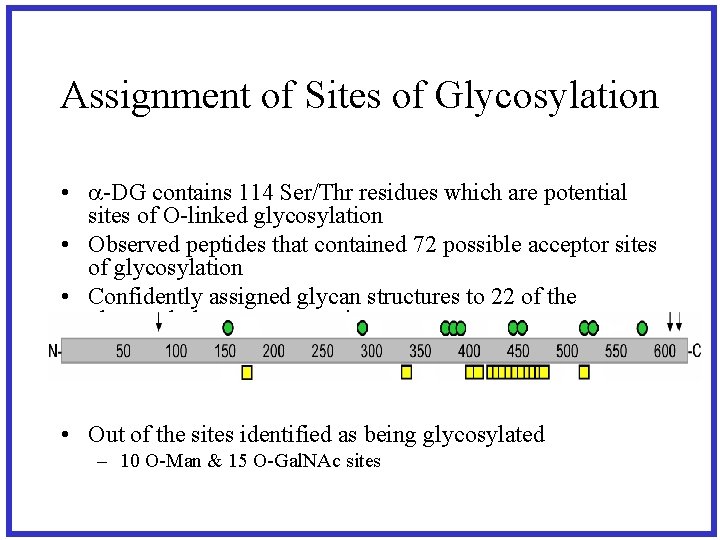
Assignment of Sites of Glycosylation • a-DG contains 114 Ser/Thr residues which are potential sites of O-linked glycosylation • Observed peptides that contained 72 possible acceptor sites of glycosylation • Confidently assigned glycan structures to 22 of the observed glycan receptor sites • Out of the sites identified as being glycosylated – 10 O-Man & 15 O-Gal. NAc sites
- Slides: 51