TANDEM AFFINITY PURIFICATION TAP TAGGING What is TAP

















- Slides: 21

TANDEM AFFINITY PURIFICATION (TAP) TAGGING

What is TAP – Tagging? INTRODUCTION: ◦ Tandem affinity purification (TAP) is a purification technique for studying protein – protein interactions. It involves creating a fusion protein with a designed piece, the TAP TAG, on the end. ◦ Tandem affinity purification (TAP) tagging is a method to purify multimeric protein complexes that can be used under essentially physiological conditions.

HISTORY: ◦ TAP Tagging was invented by a research team working in the European Molecular Biology Laboratory at late 1990 s. ◦ The first large – scale application of this technique was in 2002, to develop a visual map of the interaction of more than 230 multi – protein complexes in a yeast cell by systematically tagging the TAP Tag to each protein.

BACKGROUND: ◦ Proteins rarely act alone. ◦ Comprehensive protein interaction study thus far: 1. two-hybrid systems (ex-vivo) 2. Protein chips (in-vitro) 3. GST pull downs (in-vivo) ◦ Nowadays tandem affinity purification (TAP) and mass spectrometry is used.

PROTEIN-PROTEIN INTERACTIONS: Proteins carry out tasks together with other proteins. ◦ Multi – protein complexes (assemblies) ◦ Gene regulation ◦ Biological pathways

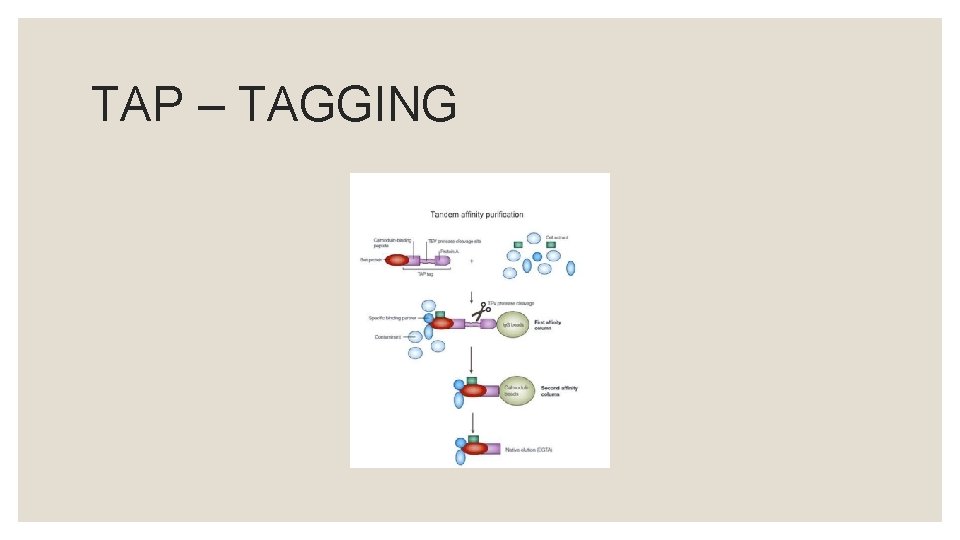
COMPONENTS OF TAP – TAG: ◦ Protein A ◦ Calmodulin Binding Peptide (CBP) ◦ Tobacco etch virus (TEV) protease cleavage site

VARIANT TAGS: ◦ This tag is also known as the C – terminal TAP Tag because an N – terminal version is also available.

TAP – TAG SELECTION AND DESIGN: Optimal tags for protein complex purification and the analysis of protein interaction should have the following characteristics: 1. High affinity for the cognate matrix for quantitative recovery of low – abundance target proteins in dilute solutions. 2. Highly specific binding to increase the ratio of specifically to non – specifically bound material to the affinity material.

TAP – TAG SELECTION AND DESIGN: 3. Efficient and specific elution allowing high – level and specific recovery of the target protein. 4. Mild conditions of elution to preserve protein interactions and protein complex structure.

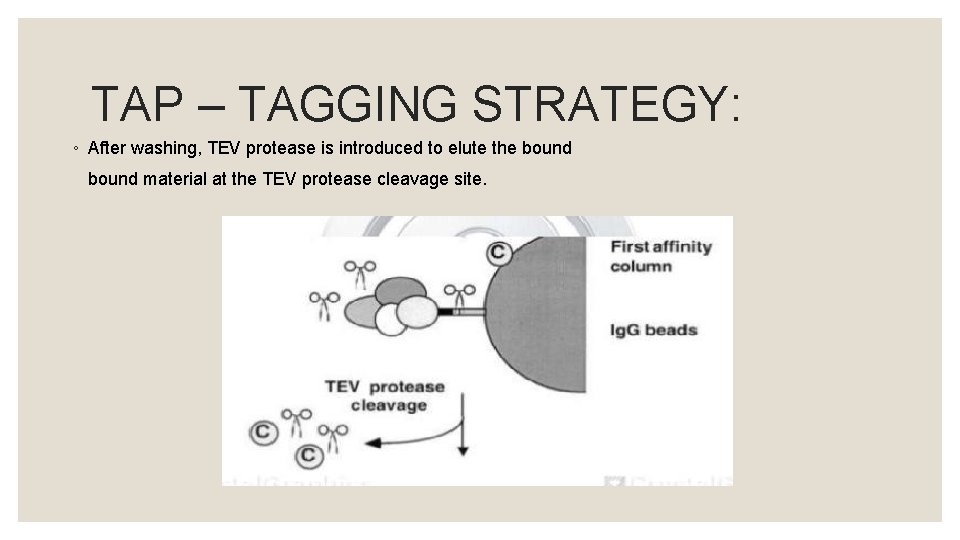
TAP – TAGGING STRATEGY: ◦ There are few methods in which the fusion protein can be introduced into the host. ◦ If the host is yeast, then plasmids will be used that will eventually translate the fusion protein within the host.

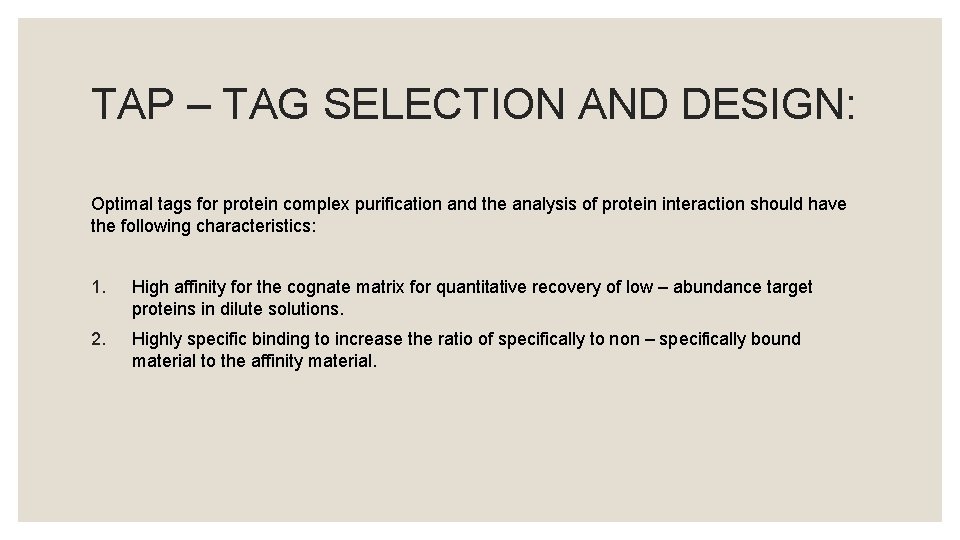
TAP – TAGGING STRATEGY: ◦ The translated fusion protein is retrieved through affinity selection, together with the other constituents attached to the new protein, by means of an Ig. G matrix.

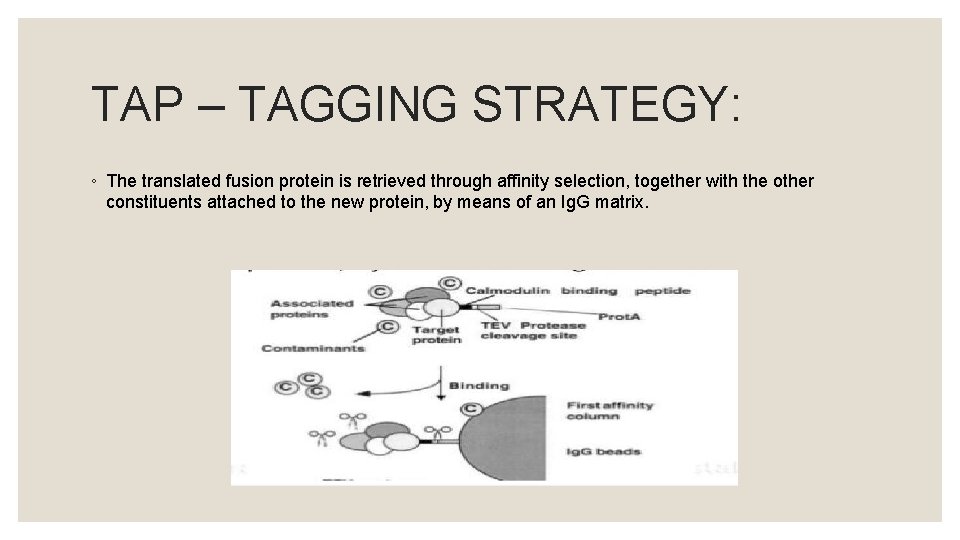
TAP – TAGGING STRATEGY: ◦ After washing, TEV protease is introduced to elute the bound material at the TEV protease cleavage site.

TAP – TAGGING STRATEGY: ◦ This eluate is then incubated with calmodulin – coated beads in the presence of calcium. ◦ This second affinity step is required to remove the TEV protease as well as traces of contaminants remaining after the first affinity step.

TAP – TAGGING STRATEGY: ◦ After washing, the eluate is then released with ethylene glycol tetra acetic acid (EGTA). SDS – PAGE or mass spectrometry can be used to analyse eluted protein.

TAP – TAGGING

ADVANTAGES: ◦ An advantage of this method is that there can be real determination of protein partners quantitatively in vivo without prior knowledge of complex composition. ◦ One of the obstacles of studying protein interaction is the contamination of the target protein especially when we don’t have any prior knowledge of it. TAP offers an effective, and highly specific means to purify target protein. After two successive affinity purifications, the chance for contaminations to be retained in the Eluate reduces significantly.

DISADVANTAGES: ◦ There is a possibility that a tag added to a protein might obscure binding of the new protein to its interacting partners. ◦ The tag may also affect protein expression levels. ◦ The tag may also not be sufficiently exposed to the affinity beads, hence skewing the results. ◦ There may also be a possibility of a cleavage of the proteins by the TEV protease.

SUITABILITY: ◦ As this method involves at leat two rounds of washing, it may not be suitable for screening transient protein interactions. ◦ But it is a good method for testing stable protein interactions by controlling the number of times the protein complex is purified.

APPLICATIONS: ◦ Tandem affinity purification (TAP) method combined with LC – MS/MS is the most accurate and reliable way to study the interaction of proteins or proteomics in a genome – wide scale. ◦ Many other proteomic analyses also involve the use of TAP tag. ◦ Researchers identified a new complex protein complex required for nuclear pre – m. RNA retention and splicing with the help of TAP – Tagging.

OTHER EPITOPE – TAG COMBINATIONS: The principle of TAP of multi proteins complexes is not limited to the combination of CBP and protein A Tags used in the original work by Rigaut et al. (1999). For example: ◦ The combination of FLAG ◦ HA – TAGS Has been used since 2000 by the group of Nakatani to purify numerous protein complexes from mammalian cells.

THANK YOU