T 2 D A complex trait Identification mapping







- Slides: 33

T 2 D- A complex trait Identification, mapping and cloning of new type 2 diabetes models from ENU mutagenesis Medical Research Council, Mammalian Genetics Unit, UK

Aim • Resolving complex genetic determinants of common disease phenotypes • ENU mutagenesis to create monogenic diabetic traits • Creation of dominant alleles of any gene that can be mutated to yield a diabetic or sub-diabetic phenotype • ENU mutagenesis to modify insulin resistance and create diabetic traits (sensitised/modifier screen) • Analysis of genes and the pathways in which they lie

Dominant monogenic ENU models



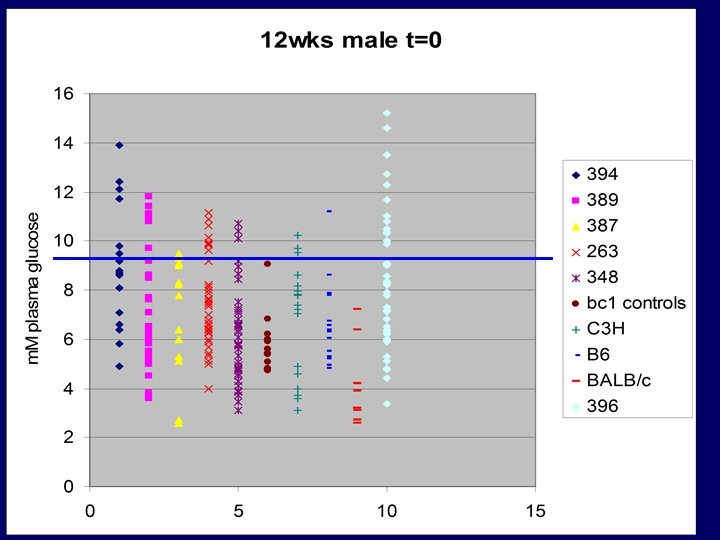
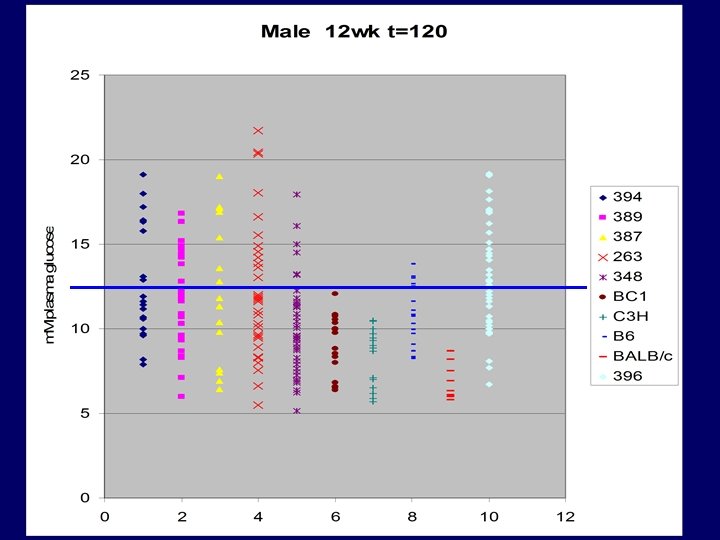
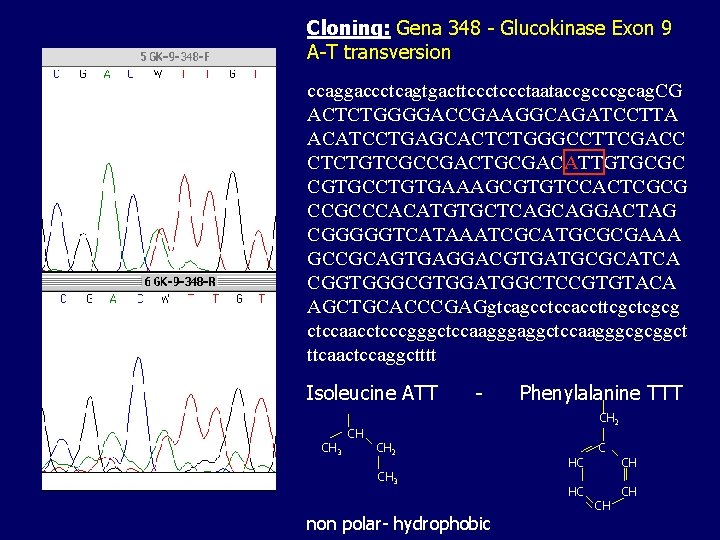
Cloning: Gena 348 - Glucokinase Exon 9 A-T transversion ccaggaccctcagtgacttccctaataccgcag. CG ACTCTGGGGACCGAAGGCAGATCCTTA ACATCCTGAGCACTCTGGGCCTTCGACC CTCTGTCGCCGACTGCGACATTGTGCGC CGTGCCTGTGAAAGCGTGTCCACTCGCG CCGCCCACATGTGCTCAGCAGGACTAG CGGGGGTCATAAATCGCATGCGCGAAA GCCGCAGTGAGGACGTGATGCGCATCA CGGTGGGCGTGGATGGCTCCGTGTACA AGCTGCACCCGAGgtcagcctccaccttcgcg ctccaacctcccgggctccaagggaggctccaagggcgcggct ttcaactccaggctttt Isoleucine ATT CH 3 CH - Phenylalanine TTT CH 2 CH 3 non polar- hydrophobic HC HC C CH CH CH

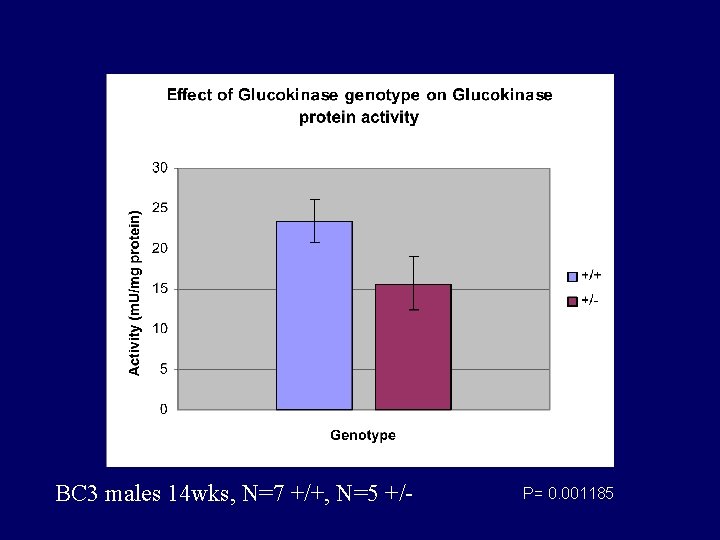
BC 3 males 14 wks, N=7 +/+, N=5 +/- P= 0. 001185

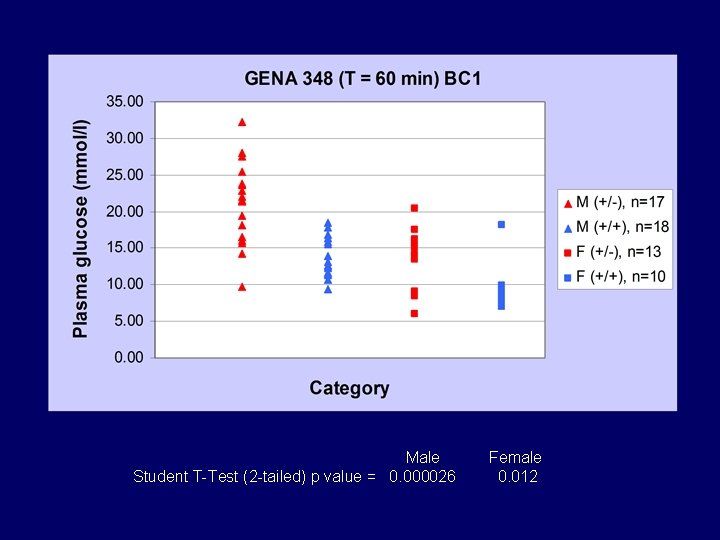
Male Student T-Test (2 -tailed) p value = 0. 000026 Female 0. 012

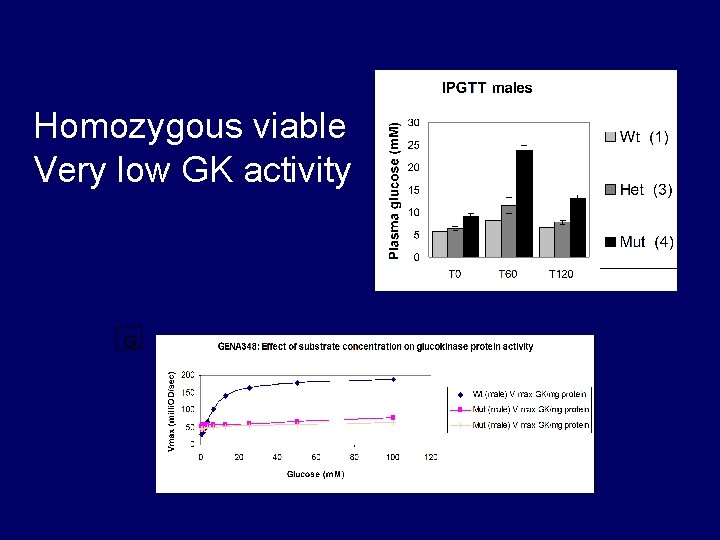
Homozygous viable Very low GK activity G

Summary: • GENA 348 GK, Isoleucine>Phenylalanine • GENA 389 Chr 11, GK splice mutation • GENA 396 Chr 11, GK splice mutation • GENA 387 Chr 11, GK splice mutation • GENA 263 Chr 6 (No GK poly) • GENA 394 Multiple linkages (No GK poly) • GENA 392 Chr 4 - Dyslipidaemia mutant

• ENU mutagenesis can be used to make new diabetic and sub-diabetic models – MODY models – Other genes • Mutations can be mapped and cloned • It should be possible to identify novel genes and/or provide new information on known diabetes genes and their pathways

Sensitised Screen Michelle Goldsworthy

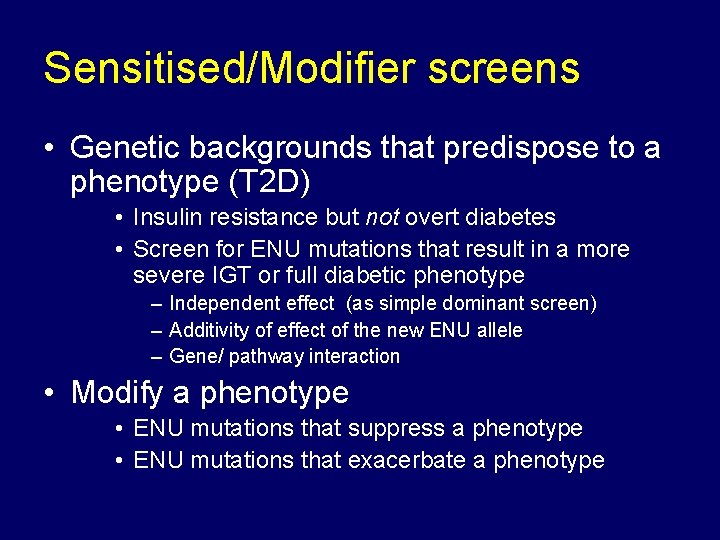
Sensitised/Modifier screens • Genetic backgrounds that predispose to a phenotype (T 2 D) • Insulin resistance but not overt diabetes • Screen for ENU mutations that result in a more severe IGT or full diabetic phenotype – Independent effect (as simple dominant screen) – Additivity of effect of the new ENU allele – Gene/ pathway interaction • Modify a phenotype • ENU mutations that suppress a phenotype • ENU mutations that exacerbate a phenotype

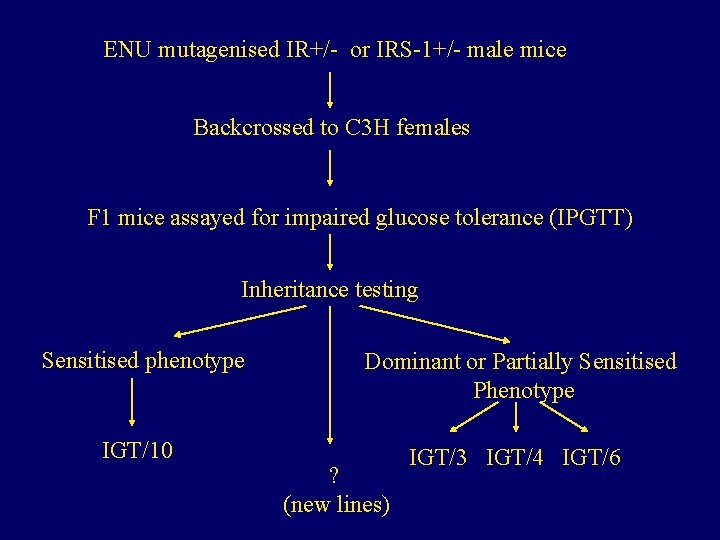
ENU mutagenised IR+/- or IRS-1+/- male mice Backcrossed to C 3 H females F 1 mice assayed for impaired glucose tolerance (IPGTT) Inheritance testing Sensitised phenotype Dominant or Partially Sensitised Phenotype IGT/10 IGT/3 IGT/4 IGT/6 ? (new lines)

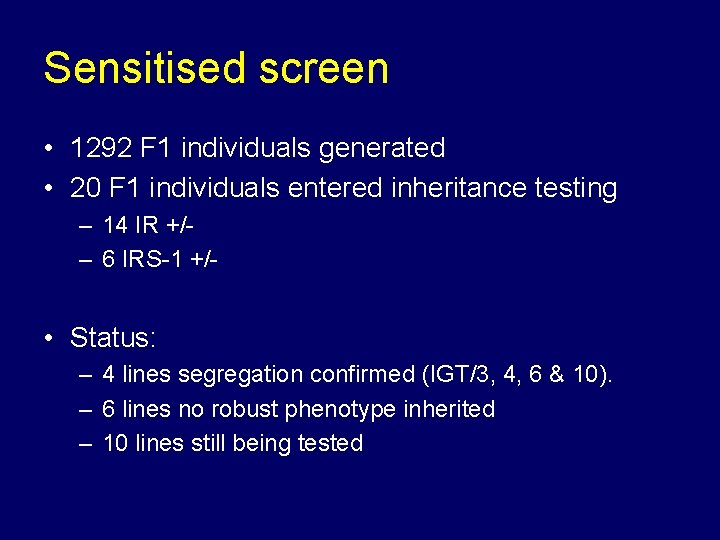
Sensitised screen • 1292 F 1 individuals generated • 20 F 1 individuals entered inheritance testing – 14 IR +/– 6 IRS-1 +/- • Status: – 4 lines segregation confirmed (IGT/3, 4, 6 & 10). – 6 lines no robust phenotype inherited – 10 lines still being tested


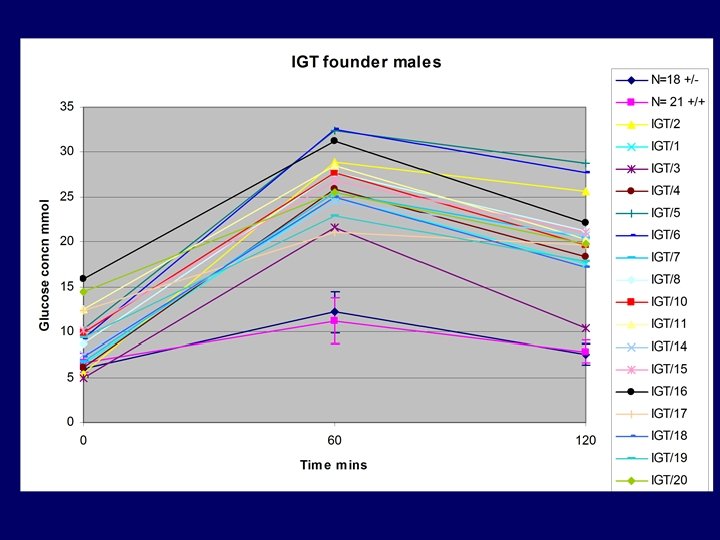
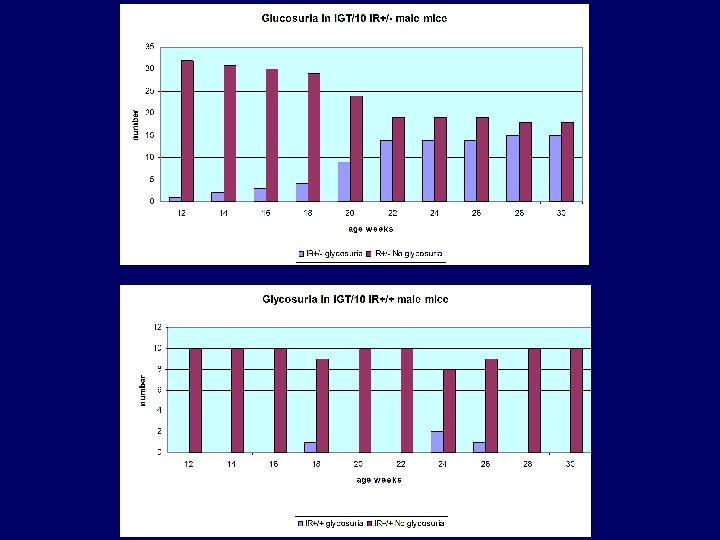
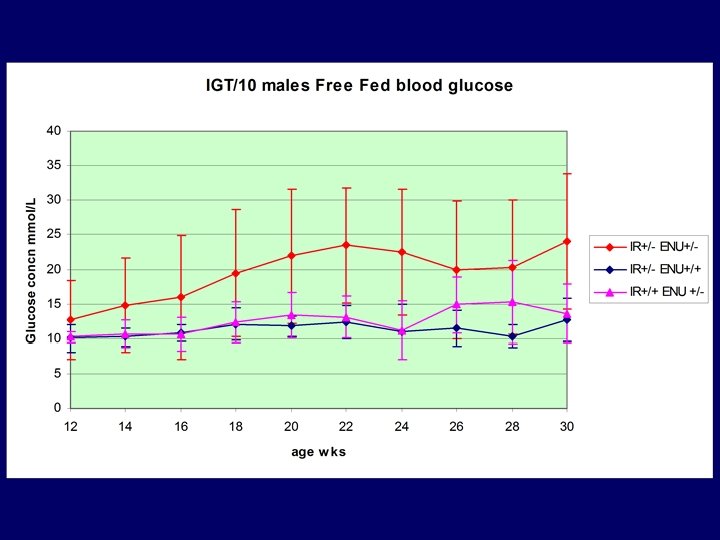
IGT/10 • 70 BC 1 mice generated – Mixture of assays – Male segregating glycosuria • 43 males and 27 females: BC 2 mice – testing at 2 weekly intervals – Glycosuria segregates in males only – Peristant glycosuria present in IR +/- only


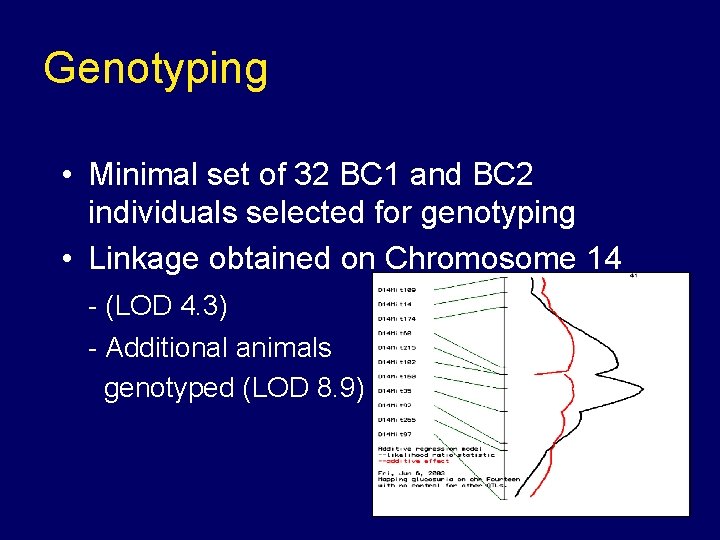
Genotyping • Minimal set of 32 BC 1 and BC 2 individuals selected for genotyping • Linkage obtained on Chromosome 14 - (LOD 4. 3) - Additional animals genotyped (LOD 8. 9)


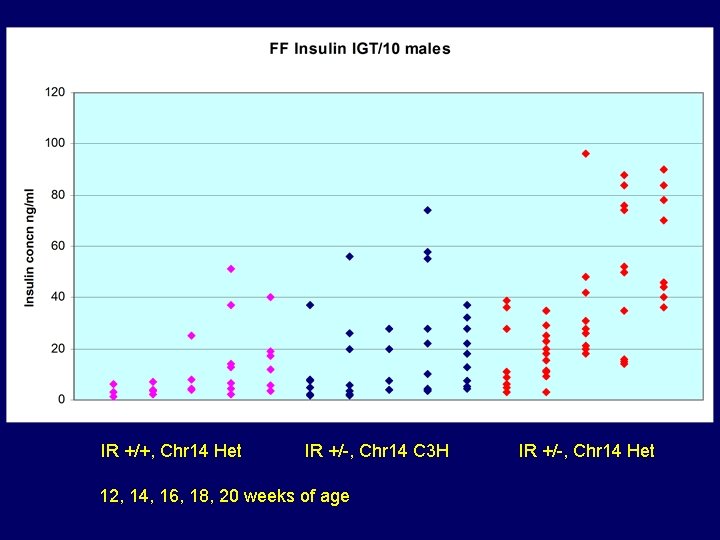
IR +/+, Chr 14 Het IR +/-, Chr 14 C 3 H 12, 14, 16, 18, 20 weeks of age IR +/-, Chr 14 Het

Future Work • Fine Mapping (Further Backcrossing) • Candidate gene sequencing • Full pathology – 9 mice (3 each group) now being analysed • Clamps • Model of Diabetic nephropathy? – Founder male shows an increase in mesangial matrix area (63. 6% vs 52. 7% P=0. 00007)

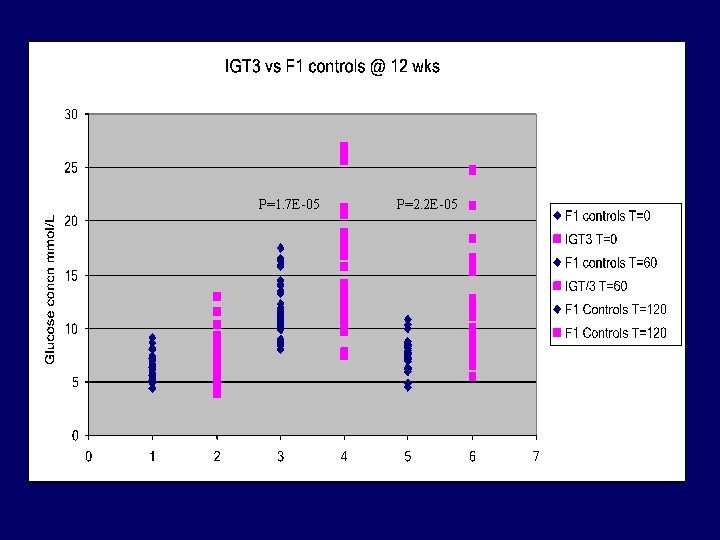
P=1. 7 E-05 P=2. 2 E-05


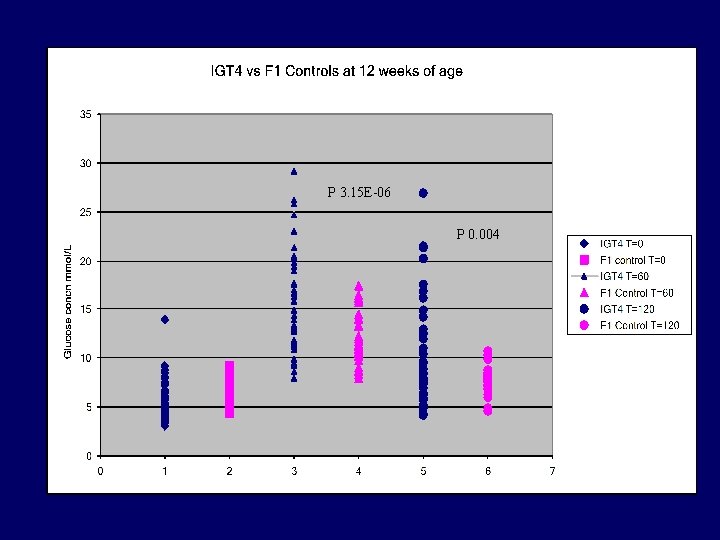
P 3. 15 E-06 P 0. 004

IGT/4 summary • Impaired glucose tolerance segregating in BC 1 and BC 2 populations – 50% IR+/- exhibit IGT – 25% IR+/+ exhibit IGT • Genotyping completed – Linkage chromosome 13 (BC 1) – Additional fine mapping underway BC 2 and BC 3 individuals (E. H) – Candidate genes (E. H)

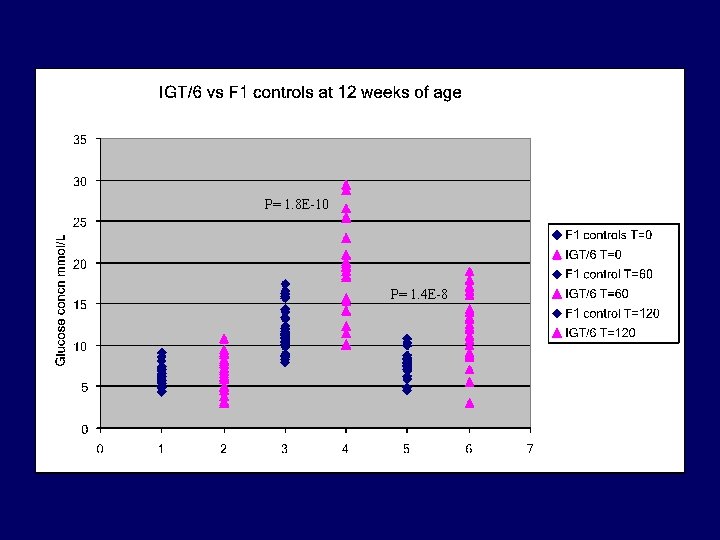
P= 1. 8 E-10 P= 1. 4 E-8

IGT/6 Summary • • Inherited impaired glucose tolerance Significantly heavier than controls Genotyping commenced BC 1 individuals (Q. A) Histological analysis - Islet cell hyperplasia - Nonalcoholic Steatohepatitis (N. A. S. H) (Q. A)

Future work • Genotyping - IGT/3 (A. H) - IGT/6 (Q. A) • Fine mapping/candidate gene analysis - IGT/4 (E. H, M. G) IGT/10 (M. G) • Histology/microarray - Islet hyperplasia IGT/3, IGT/6 (A. H, V. M) - Kidney IGT/10 (C. P) - NASH (Liver) IGT/6 (Q. A) • New Lines (J. Q, M. G)

Prospects- complementary approaches to complex traits • Dissect diseases with a complex genetic component into genes and pathways using mutagenesis approaches • Random mutagenesis may yield new genes and pathways • Sensitised screens may allow particular pathways or disease processes to be targetted

Further Prospects • Generation of allelic series in genes of interest • Reconstitution of defined polygenic models • Combination of phenotypes i. e. metabolic syndrome models • Study of disease complications • Diabetic kidney • Diabetic heart

Acknowledgements Toni Bell Jo Clay Debbie Ritson Jo Madge Lee Moir Liz Bentley Quentin Anstee (IC London) Catriona Paul Deen Quwailid Emma Horner (Etiologics) Siobhan Mansell Vesna Mijat Alison Haynes Charlotte East Alison Hugill Julie Quaterman Ayo Toye Medical Research Council Roger Cox
