Supplemental Figures APR 7 2017 Supplemental Figure 1











- Slides: 16

Supplemental Figures APR 7, 2017

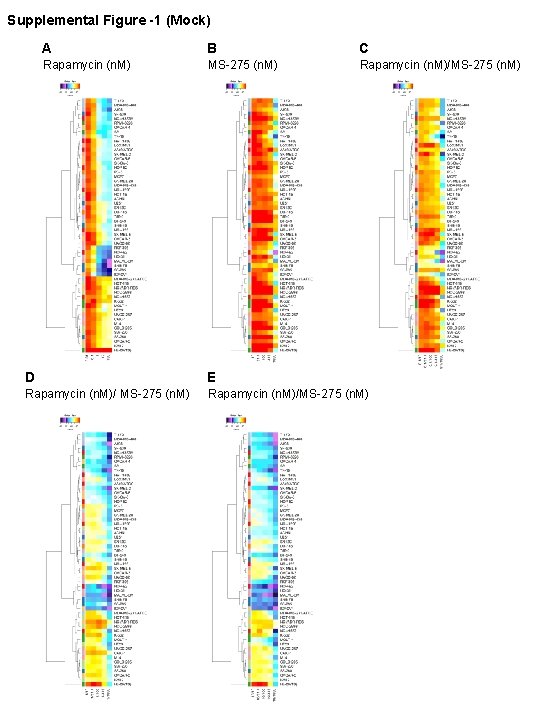
Supplemental Figure -1 (Mock) A B C Rapamycin (n. M) MS-275 (n. M) Rapamycin (n. M)/MS-275 (n. M) D E Rapamycin (n. M)/ MS-275 (n. M) Rapamycin (n. M)/MS-275 (n. M)

Figure S 1. In vitro analysis of m. TORi/HDACi activity and synergy in the NCI 60 cell lines. Heatmaps of cell growth in the NCI 60 panel after 48 hours of treatment with A) rapamycin dose course 0. 01 -100 n. M B) MS-275 dose course 31. 7 -1000 n. M C) 0. 1 n. M Rapamycin + MS-275 dose course D) 1 n. M Rapamycin + MS-275 dose course E) 10 n. M Rapamycin+MS-275 dose course. Please see https: //dtp. cancer. gov/discovery_development/nci-60/methodology. htm for detailed methods and calculations concerning cell growth in the NCI 60 panel.

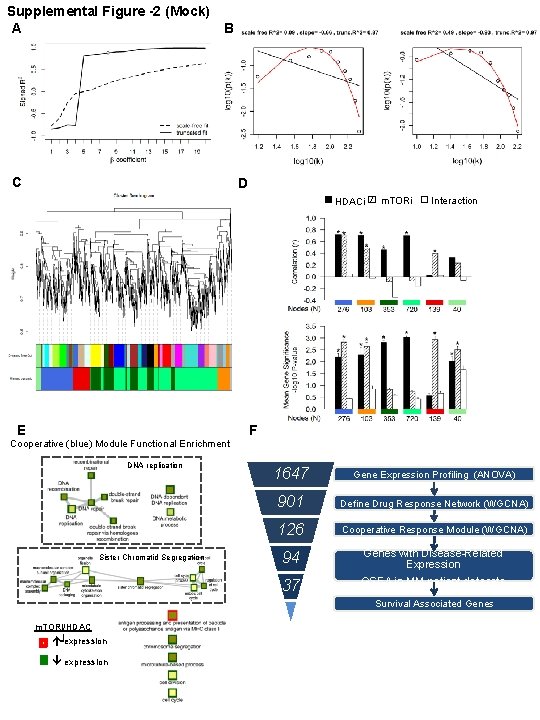
Supplemental Figure -2 (Mock) A B C D HDACi E m. TORi Interaction F Cooperative (blue) Module Functional Enrichment DNA replication Sister Chromatid Segregation 1647 Gene Expression Profiling (ANOVA) 901 Define Drug Response Network (WGCNA) 126 Cooperative Response Module (WGCNA) 94 Genes with Disease-Related Expression GSEA in MM patient datasets 37 Survival Associated Genes m. TORi/HDAC iexpression

Figure S 2. Modular network construction. A) Evaluation of the network topology models for multiple thresholds of power; a coefficient of eight was chosen as the lowest power that resulted in truncated exponential scale-free topology as measured by the fitting index (R 2 > 0. 85), B) Scale-free topology fit of the WGCNA (soft threshold = 8). The x-axis is the log 10 of connectivity (k) and the y-axis is the log 10 of the proportion of nodes with a given connectivity (p(k)). The distribution of total connectivity (left) and intra-modular connectivity (right) is shown. The black line is the power-law fit and the red line is the exponentially truncated power-law fit, C) Average hierarchical clustering dendrogram of genes using the one minus topological overalap dissimilarity metric (see Supplemental Methods). Branches of the dendrogram comprise densely interconnected, highly co-expressed genes (modules), assigned the original module colors (top bar) and the final merged module colors (bottom bar). The original modules were identified with the Dynamic Cut Tree algorithm and summarized by their first principal component of the expression values (module eigengene). Modules with highly correlated eigengenes (correlation coefficient > 0. 80) were merged into the final modules. The gray module contains the unassigned genes. D) The criteria for selection of the drug-related modules linked the ANOVA assessments of treatment effects of sirolimus and entinostat with the network module topology. On top are bar plots of the Pearson's correlation coefficients (r) of intramodular connectivity (k. IN) and gene significance (GS) values in a module, and on the bottom are bar plots of the mean gene significance in a module (MS ±SEM). An asterisk indicates that module relevance to a drug treatment was significant; both criteria were required to assign a treatment effect to the module. E) Functional enrichment of genes cooperatively regulated by m. TORi/HDACi using REVIGO visualization of functionally-related GO terms for the 126 genes in the cooperative (blue) module (depicted in Fig. 1 F, also see Table S 1 C). F) Strategy for filtering gene sets affected by the drugs. Initially 1647 genes were identified by ANOVA of microarray data as differentially expressed. Of those, WGCNA created a drug response network of 901 significantly affected genes, and further refined the cooperative drug response to 126 genes. GSEA identified 54 genes enriched in myeloma vs. healthy tissues, and 37 genes were associated with survival in multivariate survival risk predictor analyses.

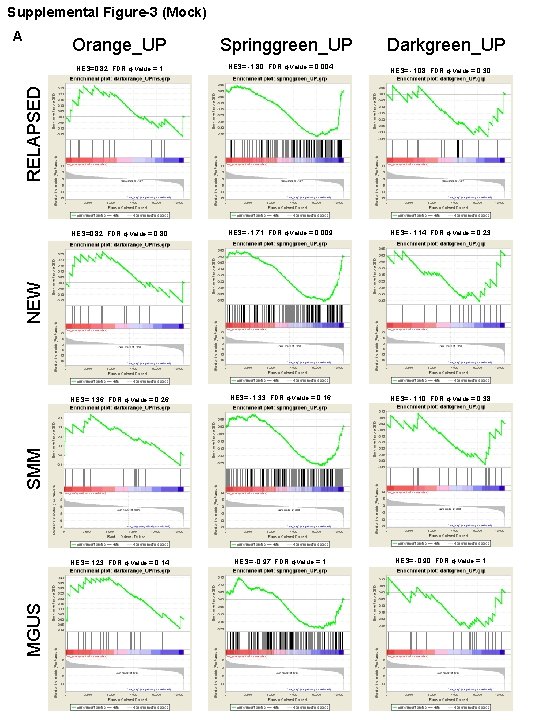
Supplemental Figure-3 (Mock) A Orange_UP Springgreen_UP Darkgreen_UP NES= -1. 80, FDR q-value = 0. 004 NES= -1. 08, FDR q-value = 0. 30 NES=0. 82, FDR q-value = 0. 80 NES= -1. 71, FDR q-value = 0. 009 NES= -1. 14, FDR q-value = 0. 23 NES= 1. 36, FDR q-value = 0. 26 NES= -1. 33, FDR q-value = 0. 16 NES= -1. 10, FDR q-value = 0. 38 NES= 1. 23, FDR q-value = 0. 14 NES= -0. 97, FDR q-value = 1 NES= -0. 90, FDR q-value = 1 MGUS SMM NEW RELAPSED NES=0. 82, FDR q-value = 1

Supplemental Figure-3 (Mock) B Springgreen_DOWNDarkgreen_DOWN Red_DOWN NES= 2. 20, FDR q-value < 1 e-4 NES= 1. 55, FDR q-value = 0. 03 NES= 0. 83, FDR q-value = 0. 99 NES= 2. 07, FDR q-value = 0. 0007 NES= 1. 72, FDR q-value = 0. 01 NES= 0. 57, FDR q-value = 0. 99 NES= 1. 11, FDR q-value = 0. 54 NES= 1. 09, FDR q-value = 0. 40 NES= -0. 61, FDR q-value = 0. 98 NES= -0. 66, FDR q-value = 1 NES= -0. 71, FDR q-value = 1 MGUS SMM NEW RELAPSED NES= 0. 65, FDR q-value = 0. 97

Figure S 3. Enrichment of drug-specific co-expression modules in disease datasets for multiple myeloma. Gene set enrichment analysis (GSEA) of the A) up-and B) downregulated genes from the modules representative of genes affected by the drug combination and individual treatments. The results for the cooperative blue module are shown in the main Figure 2 C. Module gene sets (Red_UP and Orange_DOWN) with less than 15 genes were excluded from the GSEA analysis. One-way ANOVA contrast t-statistics were used to rank the genes according to their correlation with either the Multiple Myeloma patient phenotype (red bar) or the healthy donor phenotype (blue bar). The graph on the bottom of each panel represents the ranked, ordered list of ~14, 000 unique genes. Black vertical lines show the position of individual genes from a gene set module in the ordered list of genes. The green line is the profile of the running sum of the weighted enrichment score with the maximum deviation from zero encountered in the random walk (ES). The normalized enrichment score (NES) is the enrichment score adjusted for variation in the gene set size. GSEA was performed for the four groups of multiple myeloma patients reported in GSE 6477.

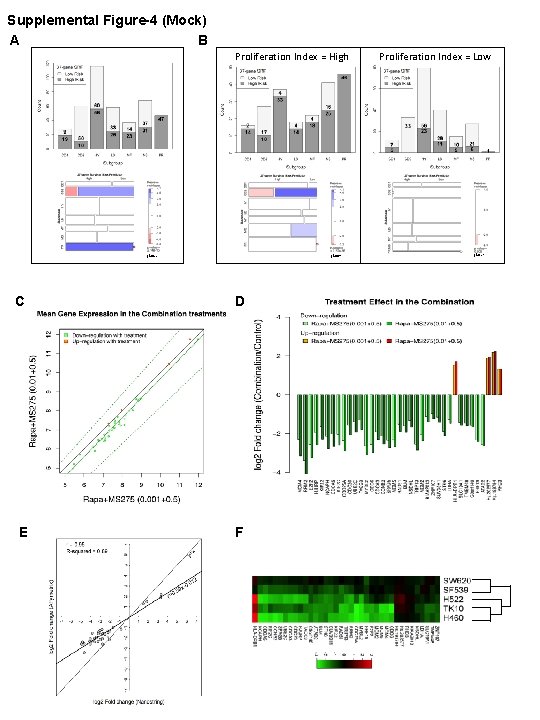
Supplemental Figure-4 (Mock) A B Proliferation Index = High c 2=81 c 2=49 C D E F Proliferation Index = Low c 2=17

Figure S 4. Relationship of 37 -gene prognostic classification with another MM classifier and pharmacodynamic effects on gene expression. A) Barplot (top) illustrating the distribution of patient groups, classified by the 37 -gene m. TORi/HDACi signature in the seven molecular subtypes of MM (CD-1, CD-2 (CCND 1/CCND 3 subgroups 1 and 2), HY (hyperdiploid), LB (low bone disease), MF (MAF/MAFB), MS (MMSET), PR (proliferation subgroup)) as defined in GSE 4581. Mosaic plot (bottom) illustrating the contingency table for the number of MM patients classified by both classifiers. In the mosaic plot each rectangle is proportional to the observed count and shaded if the residuals are significantly larger than expected by chance alone (> 2 or < -2); blue/red indicates frequencies higher/lower than expected randomly. Chi-square test was used to determine the significance for independence between the two classifications. R vcd package was used to calculate the independence test and generate the plots (Supplemental Methods). B) Barplots (top) illustrating the distribution of patient groups, classified by the 37 -gene m. TORi/HDACi signature in the seven molecular subtypes of MM (CD-1, CD-2 (CCND 1/CCND 3 subgroups 1 and 2), HY (hyperdiploid), LB (low bone disease), MF (MAF/MAFB), MS (MMSET), PR (proliferation subgroup)) as defined in GSE 4581, stratified by the Proliferation Index (PI). PI was determined for each patient as the average expression of the 11 proliferation index (PI genes). Patients with PI higher/lower than the median were classified to the High/Low stratum, respectively. Mosaic plots (bottom) illustrating the contingency table for the number of MM patients classified by the two classifiers in the High and Low PI strata. In the mosaic plots each rectangle is proportional to the observed count and shaded if the residuals are significantly larger than expected by chance alone (> 2 or < -2); blue/red indicates frequencies higher/lower than expected randomly. Since some of the expected frequencies were smaller than 5, chi-square test with permutation p-value was used to determine the significance for independence between the two classifications (5000 permutations were run). R vcd package was used to calculate the independence tests and generate the plots C-D) Two different doses of rapamycin with the same dose of MS-275 compared. Passing-Bablok linear regression analysis of the drug dose effect on L 363 transcriptional profiles of the 37 genes linked to a survival signature; two different concentrations of rapamycin (1 or 10 n. M) were compared when given in combination with 0. 5 m. M MS-275. C) Regression of the mean expression values for the 37 genes in the survival signature from GEP experiments between the two different concentrations of rapamycin (1 or 10 n. M). The correlation (Pearson’s r = 0. 988) between the two drug concentrations for expression of the 37 genes was significant (p< 2. 2 e-16). D) The treatment effect of the two different drug combinations is depicted as a log 2 fold change in gene expression of the combination treatment versus untreated L 363 cells. Gene order on the x-axis is determined by the degree of difference in gene expression fold change between the two rapamycin doses. E) Graph depicting the high correlation between expression fold change detection in combination treated L 363 cells between the Affymetrix microarray platform and the Nanostring probe-based gene expression platform. F) Heatmaps of log 2 fold change expression in the cooperative signature compared to respective untreated control samples measured by Nanostring in five NCI 60 lines: lung H 460, H 522; renal cell carcinoma TK 10; glioblastoma SF 539 and colon SW 620.

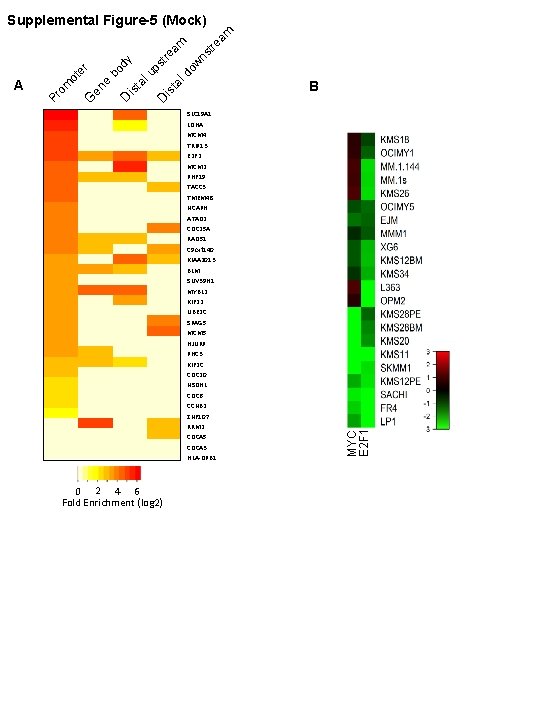
am tre ns do w ta l bo dy is ta lu ps D tre is B SLC 19 A 1 LDHA MCM 4 TRIP 13 E 2 F 2 MCM 2 PHF 19 TACC 3 TMEM 48 NCAPH ATAD 2 CDC 25 A RAD 51 C 9 orf 140 KIAA 2013 BLM SUV 39 H 1 MYBL 2 KIF 22 UBE 2 C SPAG 5 MCM 5 HJURP PHC 3 KIF 2 C CDC 20 NSDHL CDC 6 CCNB 2 ZNF 107 RRM 2 CDCA 5 CDCA 3 HLA-DPB 1 0 2 4 6 Fold Enrichment (log 2) MYC E 2 F 1 D e en G om Pr A ot er am Supplemental Figure-5 (Mock)

Figure S 5. Heatmap of the fold enrichment of c. Myc Ch. IP-seq and MYC vs E 2 F in MM Cell lines. Ch. IP signal versus input signal at each region (promoter, gene body, distal upstream, and distal downstream) of genes evaluated in the multiple myleoma MM 1 s cell line (promoter region only is shown in Figure 4 A). B) Heatmap of log 2 fold change gene expression for MYC and E 2 F 1 in the same panel of 22 myeloma cell lines from Figure 5 G.

Supplemental Figure-6 (Mock) A B C 2 -way ANOVA Myc (ON vs. OFF) Treatment Myc: Treatment Residuals Df Sum Sq Mean Sq F value Pr(>F) 1 3998. 52 85. 013 1. 22 E-08 1 7863. 81 167. 19 3. 59 E-11 1 3998. 52 85. 01 1. 22 E-08 20 940. 68 47. 03 1 -way anova contrast Estimate Std. Error t value Pr(>|t|) lower CI upper CI Class OFF. RM vs. ON. RM 51. 63 3. 96 13. 04 3. 09 E-11 43. 37 59. 89

Figure S 6. A) Graph showing the log 2 fold change in m. RNA over time of MYC and E 2 F 1 in a P 493 -6 dox-removal time course experiment. B) Viability data from P 493 -6 experiment testing drug effects (n = 2 experiments with 3 technical replicates in each) with and without MYC expression, (also shown in Figure 4 D) for which a C) 2 -way ANOVA was performed.

80 60 40 (Ch ) x 10 m A MYC Half-life Immediately Following Drug Treatment 20 in Supplemental Figure -7 (Mock) Un T 1/2=28. 0 m Rapa T 1/2=17. 7 m MS T 1/2=22. 7 m Rapa+ MS T 1/2=15. 7 m B C MYC Half-life 4 Hours Following Drug Treatment

Figure S 7. Early time point MYC stability experiments in L 363. Cycloheximide/MYC stability experiments (same design as Figure 4 E), but rapamycin and/or MS-275 treatment was begun A) immediately after 5 minute cycloheximide pre-treatment to show immediate effects of each single agent and the combination on MYC protein stability or B)cycloheximide treatment 4 hours after drug treatment. C)Table of slope-equations and half-life calculations from cycloheximide experiments examining MYC stability immediately or 18 hours after drug treatment.
 Supplementary figures
Supplementary figures Supplementary figure
Supplementary figure Supplemental figure
Supplemental figure Program
Program Sistem za centralizovano prijavljivanje korisnika apr
Sistem za centralizovano prijavljivanje korisnika apr Nominal interest rate equation
Nominal interest rate equation Annual product quality review
Annual product quality review Cours cap apr
Cours cap apr What does apr stand for
What does apr stand for Air purifying respirator (apr)
Air purifying respirator (apr) Jan, feb, mar
Jan, feb, mar Apr vs ear
Apr vs ear àpr
àpr Apr vs ear
Apr vs ear Two shapes that are similar are also congruent
Two shapes that are similar are also congruent Solid and plane figures
Solid and plane figures Plane figures
Plane figures