Summary of singlemolecule experiments Motor proteins Are unidirectional











- Slides: 36

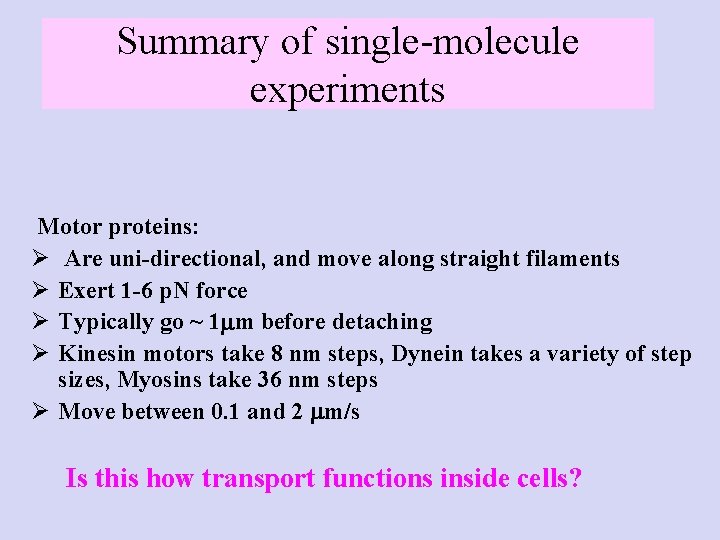
Summary of single-molecule experiments Motor proteins: Ø Are uni-directional, and move along straight filaments Ø Exert 1 -6 p. N force Ø Typically go ~ 1 m before detaching Ø Kinesin motors take 8 nm steps, Dynein takes a variety of step sizes, Myosins take 36 nm steps Ø Move between 0. 1 and 2 m/s Is this how transport functions inside cells?

How do we go from singlemolecule characterization to in vivo function?

Herpes virus in cultured neuron

Why do cargos need multiple motors? Many intercellular distances are longer than 1 micron

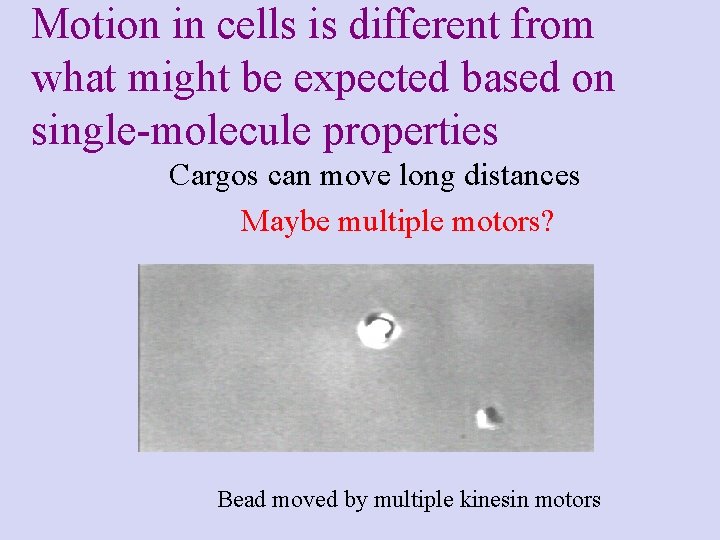
Motion in cells is different from what might be expected based on single-molecule properties Cargos can move long distances Maybe multiple motors? Bead moved by multiple kinesin motors

So, multiple motors can move a cargo long distances. Now, lets look more carefully… Start to build complexity in a controlled environment, i. e. in vitro, and understand how motors work together

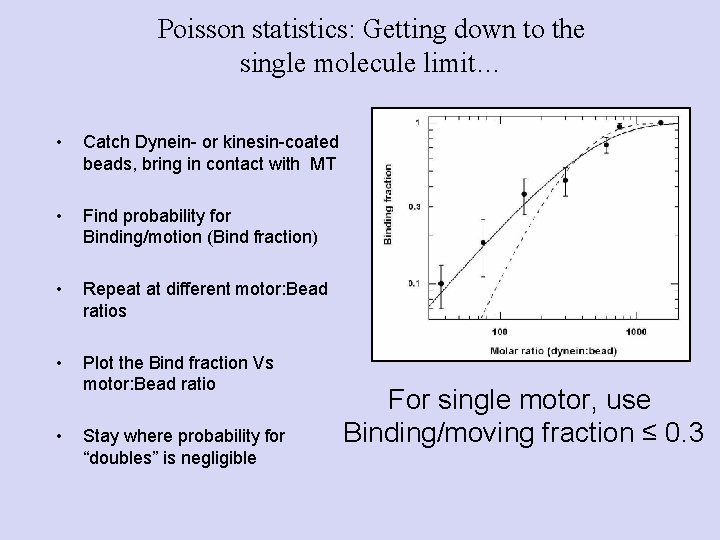
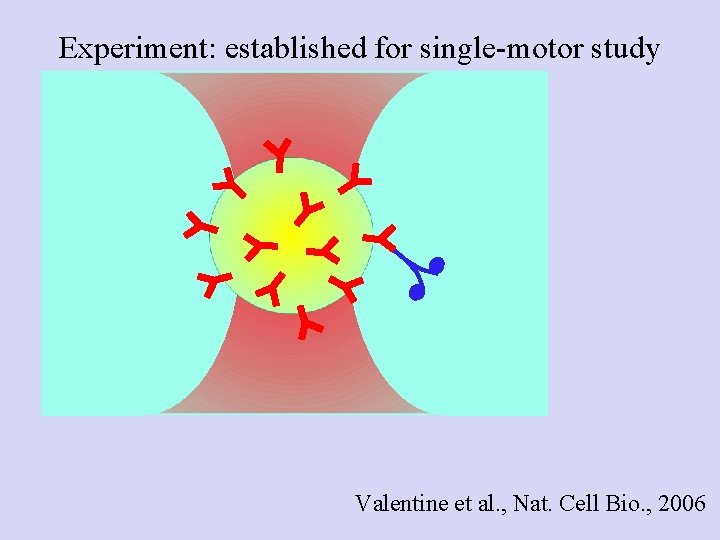
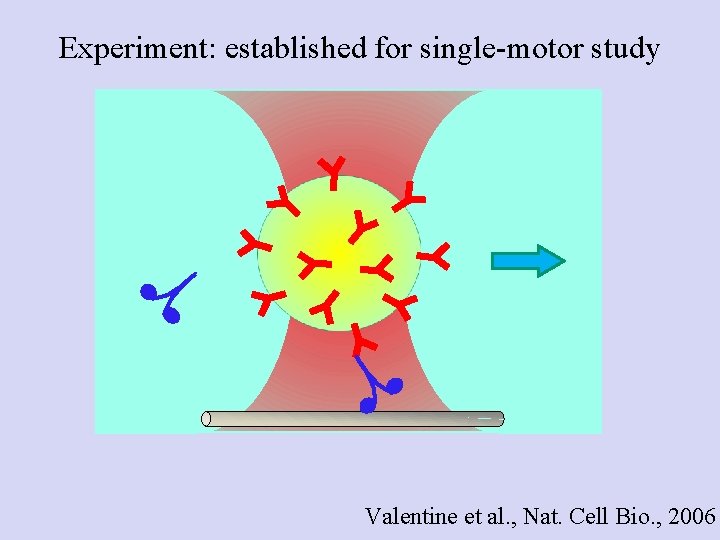
Poisson statistics: Getting down to the single molecule limit… • Catch Dynein- or kinesin-coated beads, bring in contact with MT • Find probability for Binding/motion (Bind fraction) • Repeat at different motor: Bead ratios • Plot the Bind fraction Vs motor: Bead ratio • Stay where probability for “doubles” is negligible For single motor, use Binding/moving fraction ≤ 0. 3

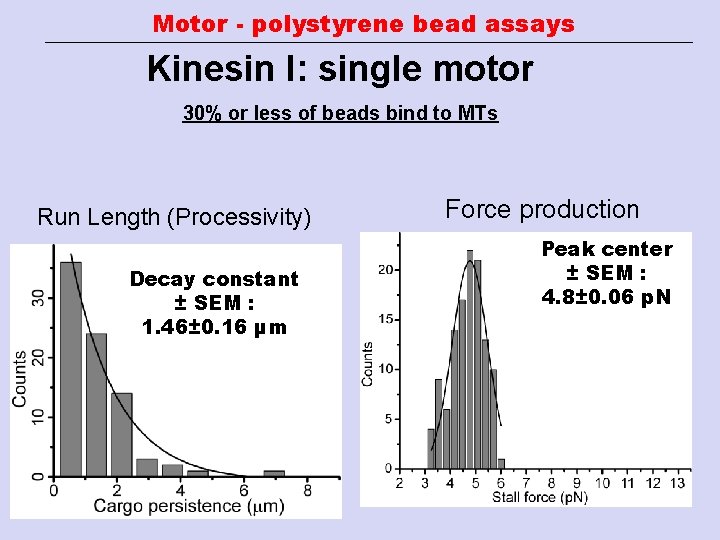
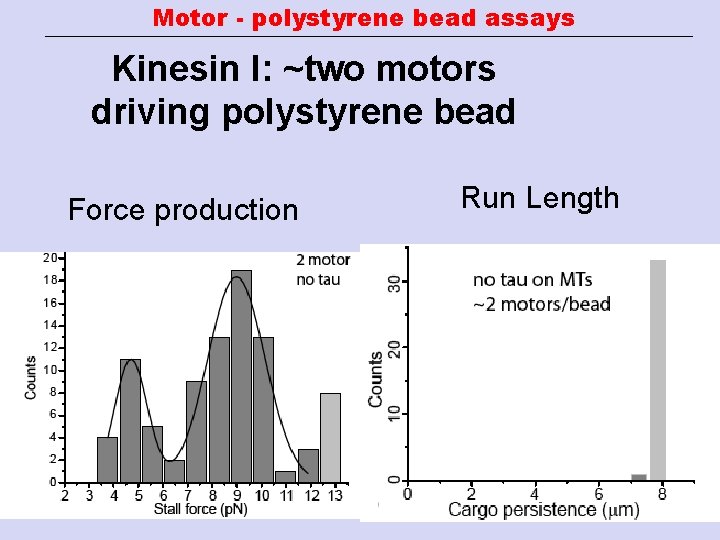
Motor - polystyrene bead assays Kinesin I: single motor 30% or less of beads bind to MTs Run Length (Processivity) Decay constant ± SEM : 1. 46± 0. 16 µm Force production Peak center ± SEM : 4. 8± 0. 06 p. N

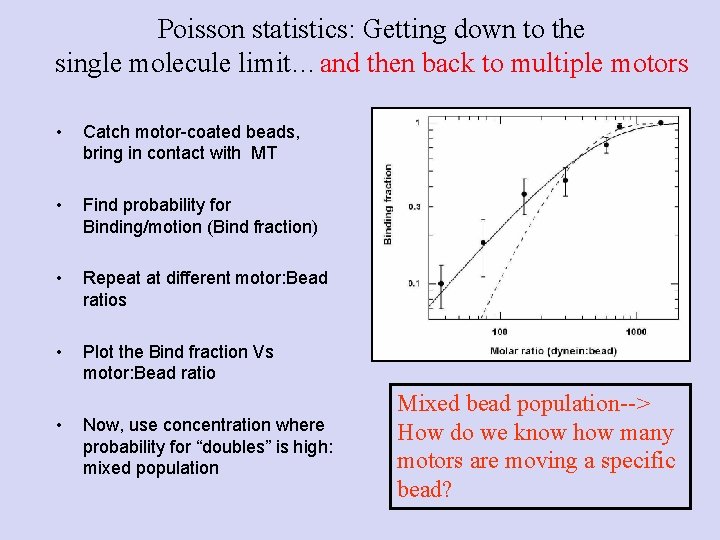
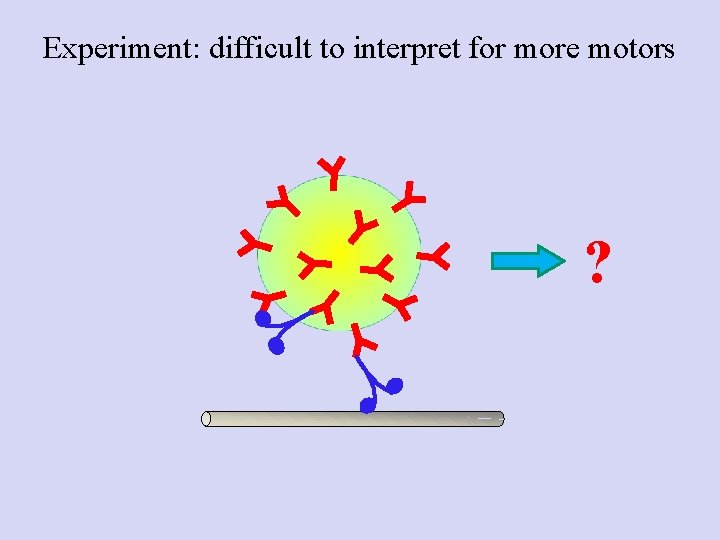
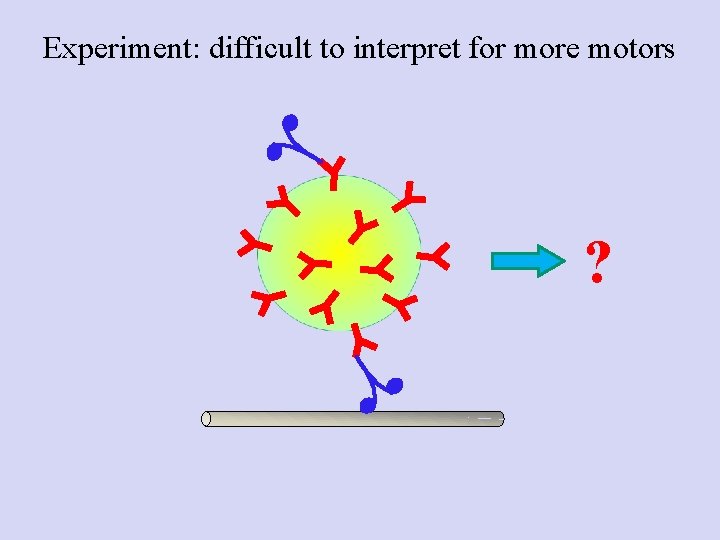
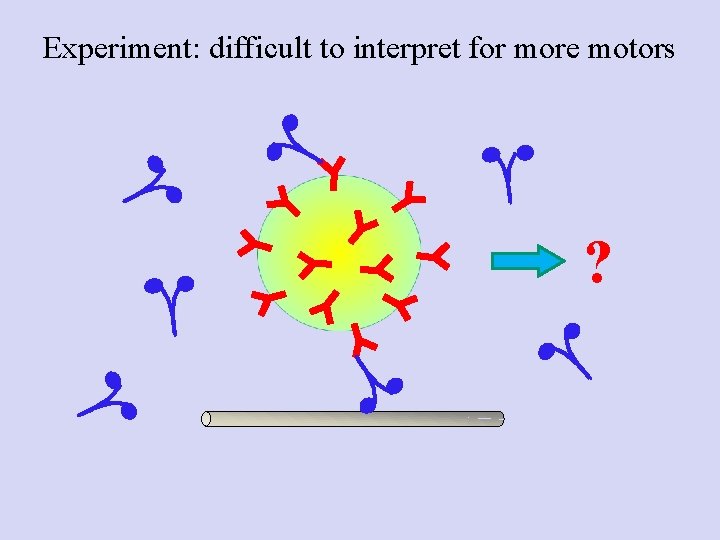
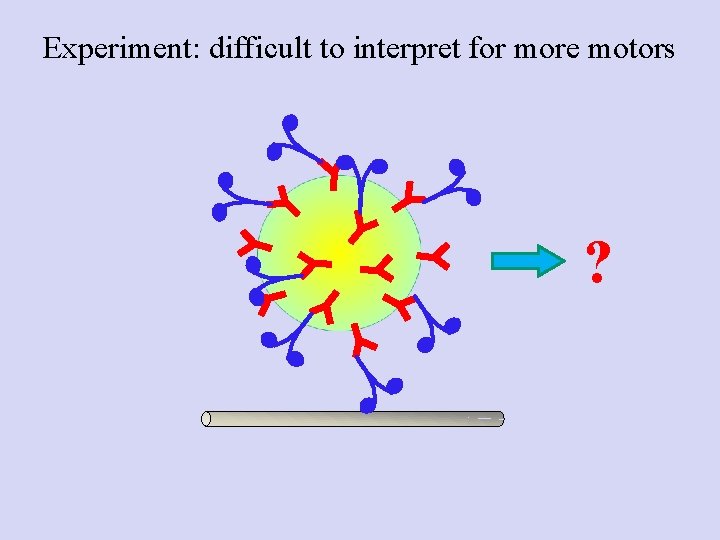
Poisson statistics: Getting down to the single molecule limit…and then back to multiple motors • Catch motor-coated beads, bring in contact with MT • Find probability for Binding/motion (Bind fraction) • Repeat at different motor: Bead ratios • Plot the Bind fraction Vs motor: Bead ratio • Now, use concentration where probability for “doubles” is high: mixed population Mixed bead population--> How do we know how many motors are moving a specific bead?

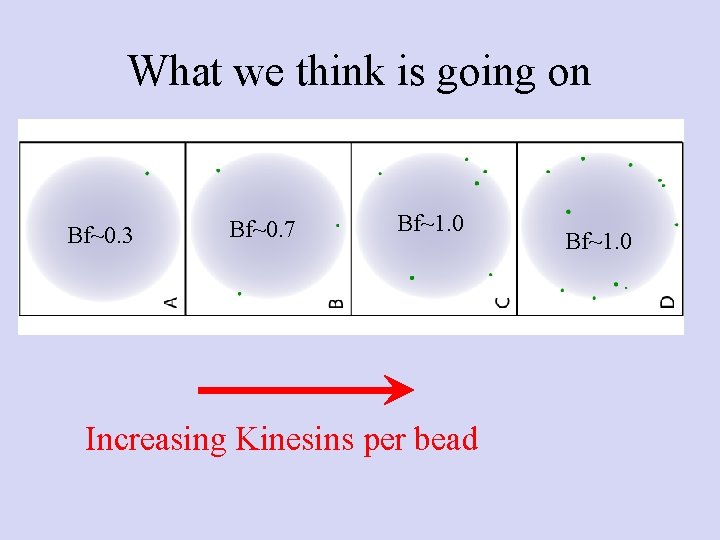
What we think is going on Bf~0. 3 Bf~0. 7 Bf~1. 0 Increasing Kinesins per bead Bf~1. 0

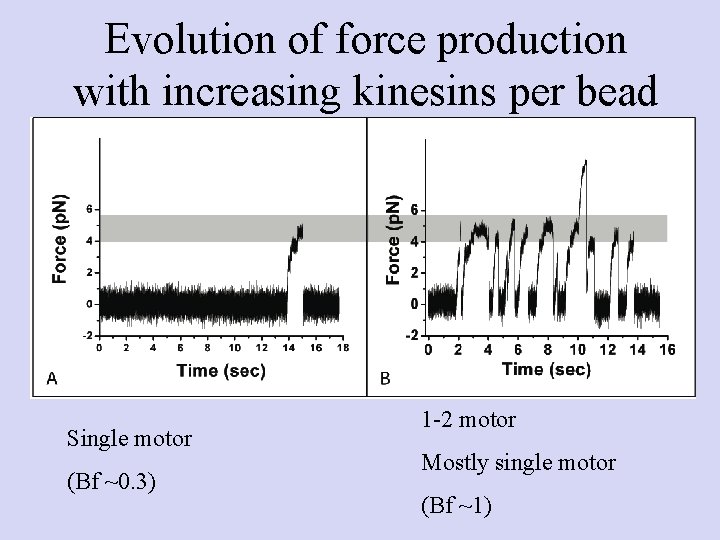
Evolution of force production with increasing kinesins per bead Single motor (Bf ~0. 3) 1 -2 motor Mostly single motor (Bf ~1)

Conclusion: for multiple-motor driven transport, binding fraction cannot tell you how many motors engaged. Stalling forces are additive at low motor number; use this as a readout of the number of instantaneously engaged motors

Motor - polystyrene bead assays Kinesin I: ~two motors driving polystyrene bead Force production Run Length

Summary for ~2 engaged Kinesins: * Velocities unchanged (not shown) * Stall forces ~ additive * Cargo travel lengths very long, but this is not really correct (see next) >> Similar results for cytoplasmic dynein (see Mallik et al, Curr. Bio, 2005) More: see website bioweb. bio. uci. edu/sgross

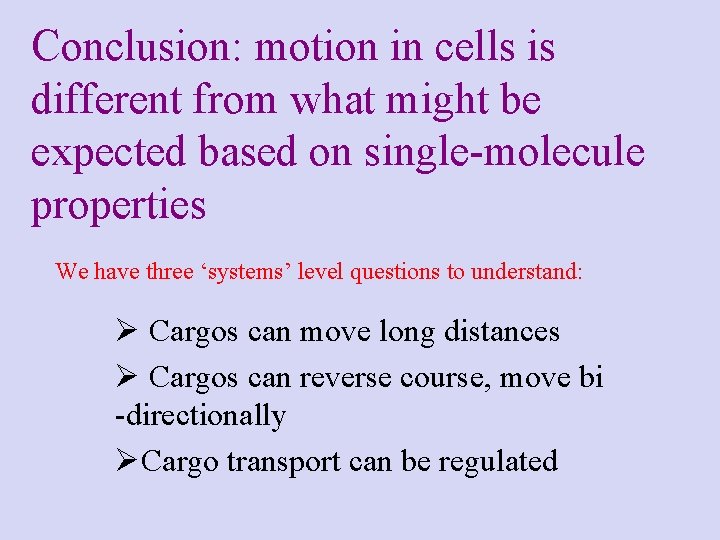
Conclusion: motion in cells is different from what might be expected based on single-molecule properties We have three ‘systems’ level questions to understand: Ø Cargos can move long distances Ø Cargos can reverse course, move bi -directionally ØCargo transport can be regulated

What single-molecule properties are particularly important for how multiple motors function together?

Cartoon of processive motion of a cargo moved by two motors

From cartoon… On-rate Off-rate Overall number of motors

Analytic Mean-field theory of average cargo travel carried by two motors p: binding rate (1/s) e: unbinding rate (1/s) d=v*(1/ e)=v/ e Velocity: crucial initial condition Klumpp and Lipowsky, PNAS, 2005 Xu et al, Traffic, 2012

Experiment: established for single-motor study Valentine et al. , Nat. Cell Bio. , 2006

Experiment: established for single-motor study Valentine et al. , Nat. Cell Bio. , 2006

Experiment: difficult to interpret for more motors ?

Experiment: difficult to interpret for more motors ?

Experiment: difficult to interpret for more motors ?

Experiment: difficult to interpret for more motors ?

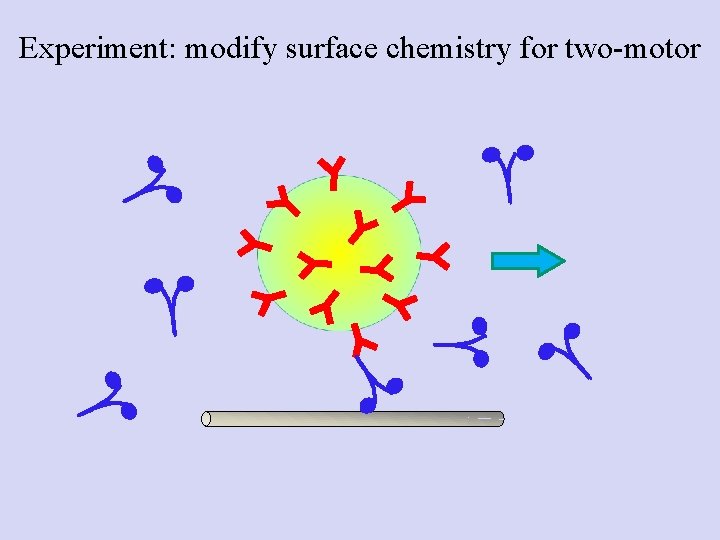
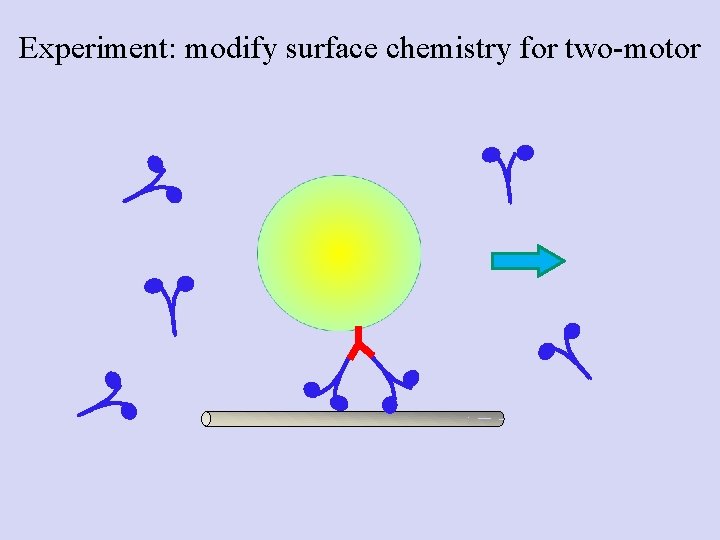
Experiment: modify surface chemistry for two-motor

Experiment: modify surface chemistry for two-motor

Position Experiment: force to further require two-motor Time

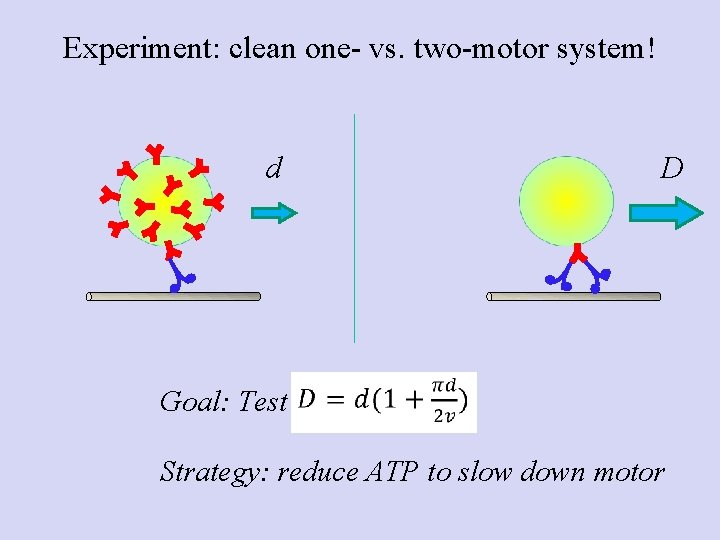
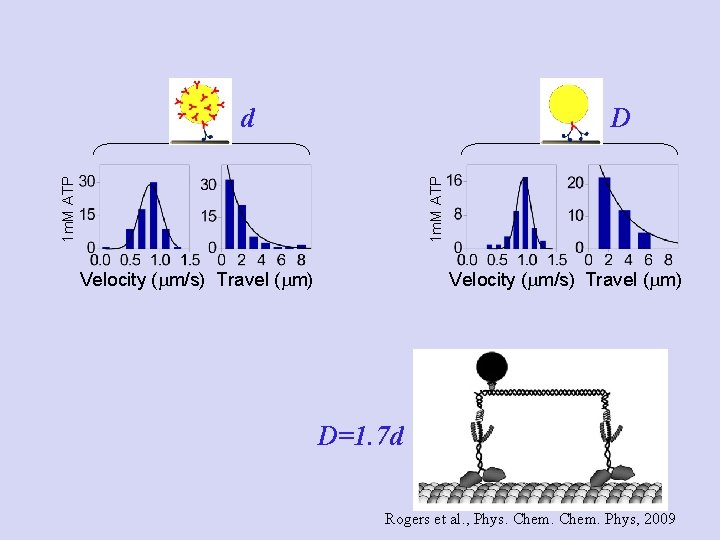
Experiment: clean one- vs. two-motor system! d D Goal: Test Strategy: reduce ATP to slow down motor

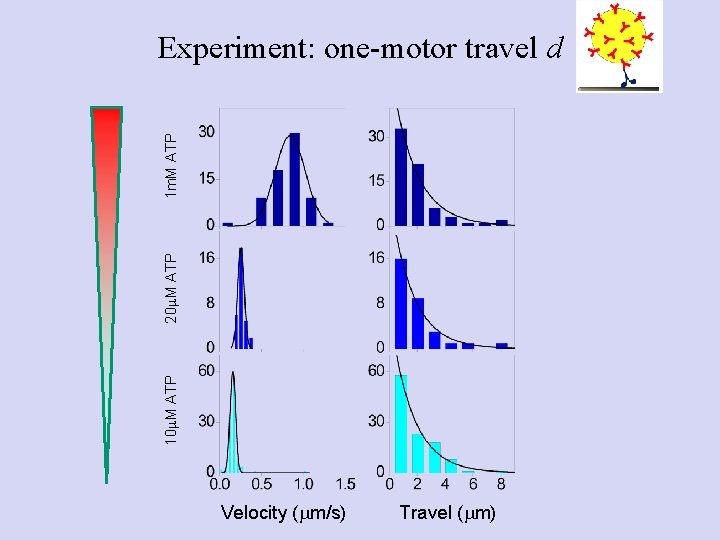
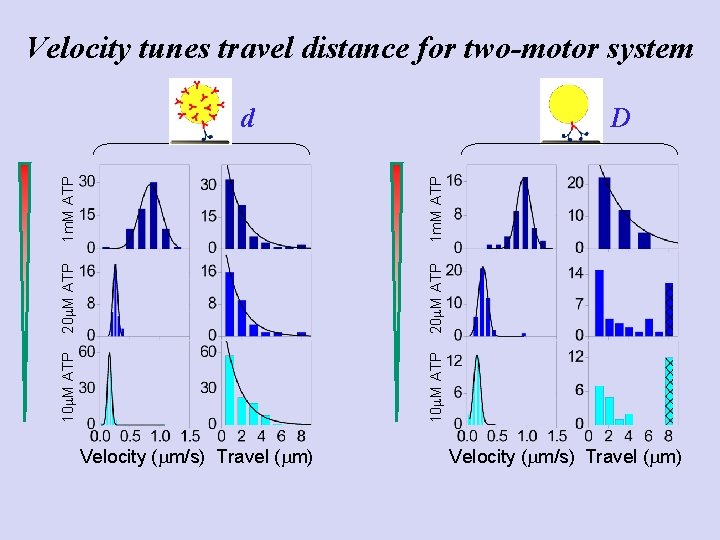
10 m. M ATP 20 m. M ATP 1 m. M ATP Experiment: one-motor travel d Velocity (mm/s) Travel (mm)

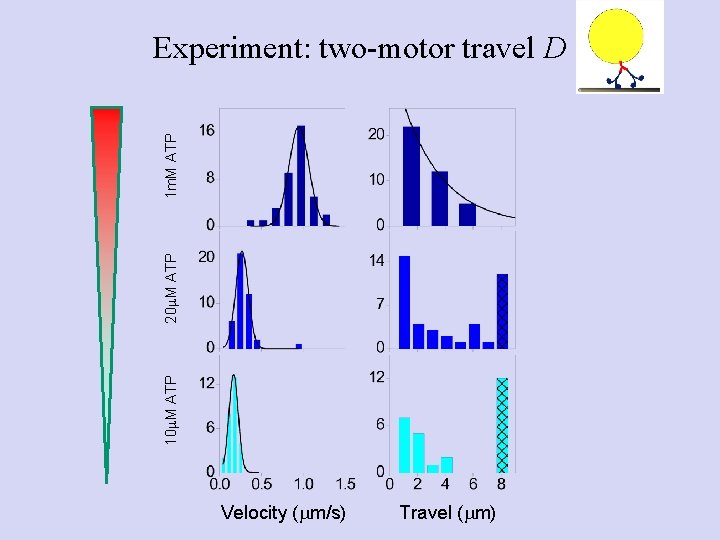
10 m. M ATP 20 m. M ATP 1 m. M ATP Experiment: two-motor travel D Velocity (mm/s) Travel (mm)

D 1 m. M ATP d Velocity (mm/s) Travel (mm) D=1. 7 d Rogers et al. , Phys. Chem. Phys, 2009

Velocity tunes travel distance for two-motor system D 10 m. M ATP 20 m. M ATP 1 m. M ATP d Velocity (mm/s) Travel (mm)

Motors work in small ensemble in cells We establish velocity as a control for ensemble travel May be particularly important, as beautiful work by Joanny (Campas, et al, Biophys. J. , 2008) suggests a limited number of motors (~ 9 max) can be active.

Hw #2 : Model two kinesin motors functioning together, and then investigate velocity effects. In Hw #1 you developed a simulation for 1 motor. Here, stick two such motors together. Assume initially that the motors here have the same properties as in the previous hw. The main goal here is to get the simulation working, and compare its results for a few different choices of ‘on’ rates and ‘off’ rates to the order-of-magnitude theory developed in class. How similar are the two sets of predictions? For a single motor with processivity of 1. 2 microns, what is your prediction for the mean travel of a cargo with two such motors, assuming an ‘on’ rate of 2/sec or 5/sec. Do this assuming a velocity of 800 nm/sec, and a velocity of 100 nm/sec.

Velocity: the link between temporal and spatial of individual motor unbinding from microtubule Slower velocity buys more time for additional motor to bind before the current bound one detaches. (see Xu et al, Traffic, 2012)
 Antigentest åre
Antigentest åre Unidirectional hydraulic motor symbol
Unidirectional hydraulic motor symbol Gear pump symbol
Gear pump symbol Section 8-1 summary carbohydrates fats and proteins
Section 8-1 summary carbohydrates fats and proteins Chapter 2 american experiments summary
Chapter 2 american experiments summary Unidirectional restrictor
Unidirectional restrictor Bis recommended orthographic projection
Bis recommended orthographic projection Dimensioning rules
Dimensioning rules Unidirectional method
Unidirectional method Control bus unidirectional or bidirectional
Control bus unidirectional or bidirectional Unidirectional system the dimensions are
Unidirectional system the dimensions are Unidirectional distribution systems
Unidirectional distribution systems In unidirectional system the dimensions are
In unidirectional system the dimensions are Hypoplexy
Hypoplexy Unidirectional flow of electric charge
Unidirectional flow of electric charge Pony motor starting method diagram
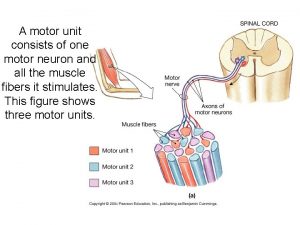
Pony motor starting method diagram A motor unit consists of:
A motor unit consists of: Hunting in electrical machines
Hunting in electrical machines Motor parts name
Motor parts name Ee 216
Ee 216 Protein pump vs protein channel
Protein pump vs protein channel Globular vs fibrous proteins
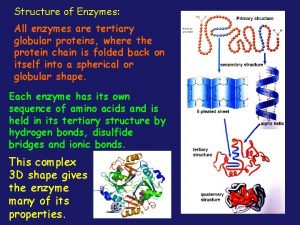
Globular vs fibrous proteins All enzymes are globular proteins
All enzymes are globular proteins Section 8-1 carbohydrates fats and proteins answer key
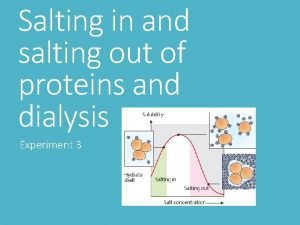
Section 8-1 carbohydrates fats and proteins answer key Salting in and salting out of proteins
Salting in and salting out of proteins Building blocks of protein
Building blocks of protein Qualitative tests for proteins
Qualitative tests for proteins Primary derived proteins
Primary derived proteins Monomers of proteins
Monomers of proteins Structural protein
Structural protein Bilirubin bruise
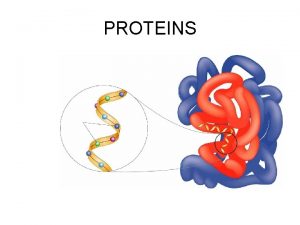
Bilirubin bruise Protein folding
Protein folding Structure database in bioinformatics
Structure database in bioinformatics Metodo di lowry
Metodo di lowry Complementary proteins
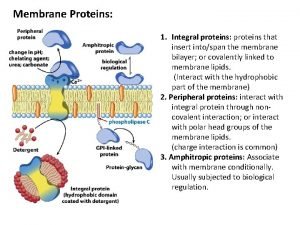
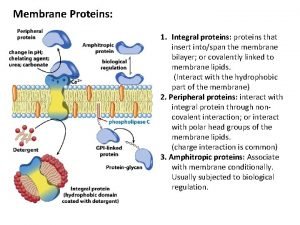
Complementary proteins Functions of membrane proteins
Functions of membrane proteins Motifs and domains of proteins
Motifs and domains of proteins