Sulfate Reducing Bacteria Global S Cycle S 0
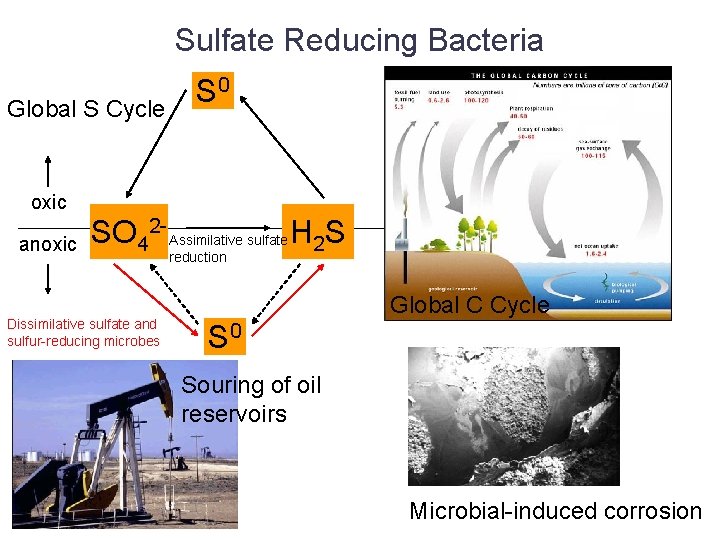
Sulfate Reducing Bacteria Global S Cycle S 0 oxic anoxic SO 42 - Assimilative sulfate H 2 S Dissimilative sulfate and sulfur-reducing microbes reduction Global C Cycle S 0 Souring of oil reservoirs Microbial-induced corrosion
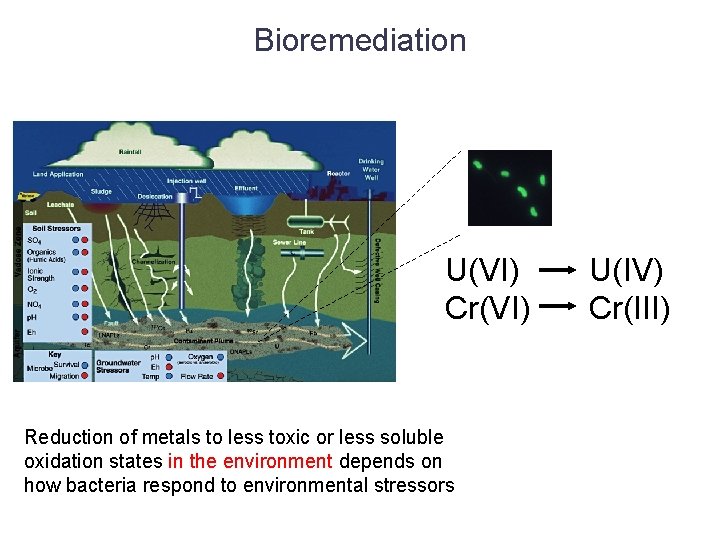
Bioremediation U(VI) Cr(VI) Reduction of metals to less toxic or less soluble oxidation states in the environment depends on how bacteria respond to environmental stressors U(IV) Cr(III)
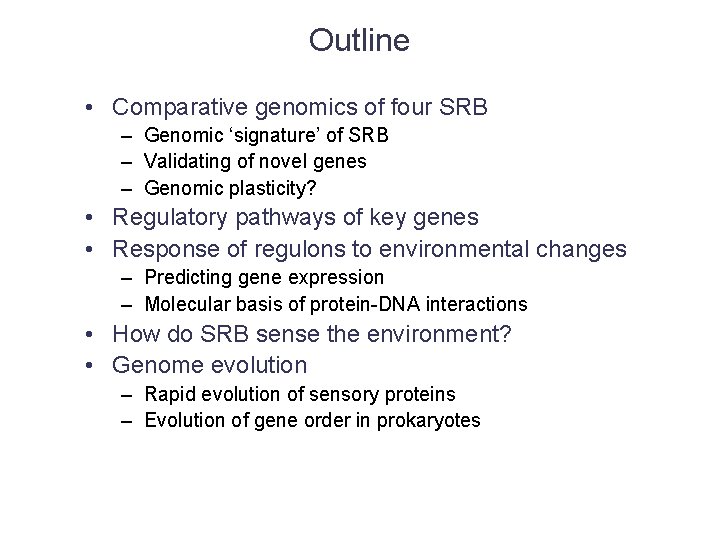
Outline • Comparative genomics of four SRB – Genomic ‘signature’ of SRB – Validating of novel genes – Genomic plasticity? • Regulatory pathways of key genes • Response of regulons to environmental changes – Predicting gene expression – Molecular basis of protein-DNA interactions • How do SRB sense the environment? • Genome evolution – Rapid evolution of sensory proteins – Evolution of gene order in prokaryotes
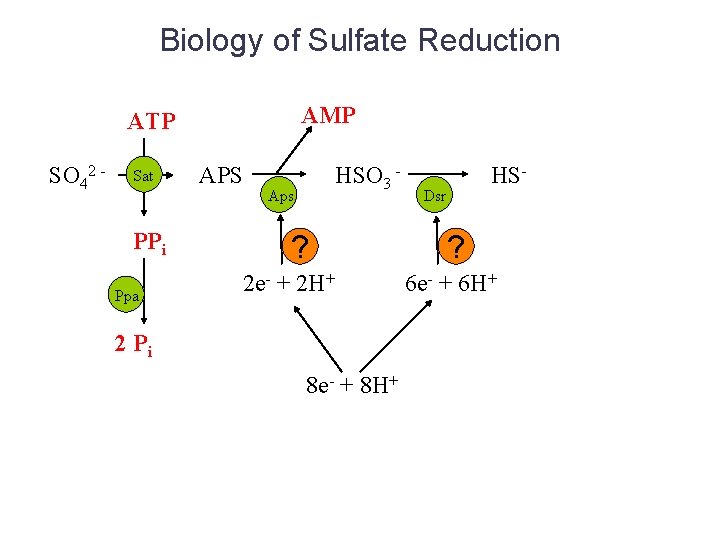
Biology of Sulfate Reduction AMP ATP SO 42 - Sat PPi Ppa APS HSO 3 - Aps ? 2 e- + 2 H+ 2 Pi 8 e- + 8 H+ HS- Dsr ? 6 e- + 6 H+

Biology of Sulfate Reduction AMP ATP SO 42 - Sat APS PPi Ppa HSO 3 - Aps ? 2 Pi 8 e- + 8 H+ 2 lactate 2 ADP + 2 Pi Dsr ? 2 e- + 2 H+ Ldh, Por Pta, Ack HS- 2 acetate + 2 CO 2 2 ATP 6 e- + 6 H+
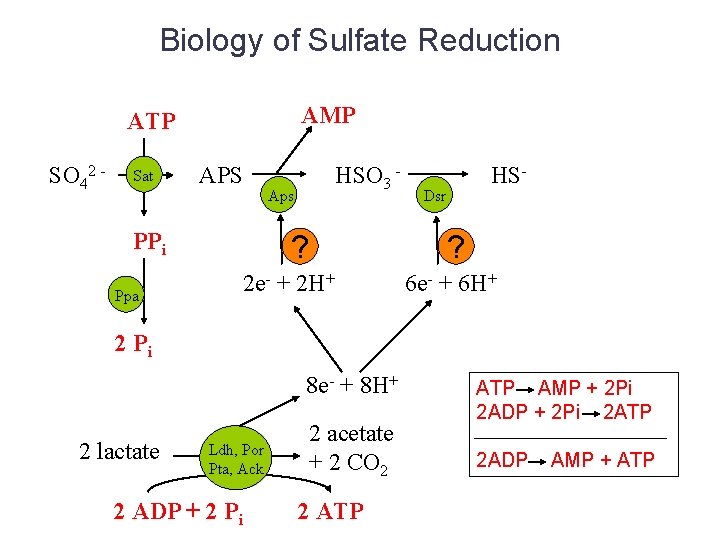
Biology of Sulfate Reduction AMP ATP SO 42 - Sat APS PPi Ppa HSO 3 - Aps ? HS- Dsr ? 2 e- + 2 H+ 6 e- + 6 H+ 2 Pi 8 e- + 8 H+ 2 lactate Ldh, Por Pta, Ack 2 ADP + 2 Pi 2 acetate + 2 CO 2 2 ATP AMP + 2 Pi 2 ADP + 2 Pi 2 ATP 2 ADP AMP + ATP
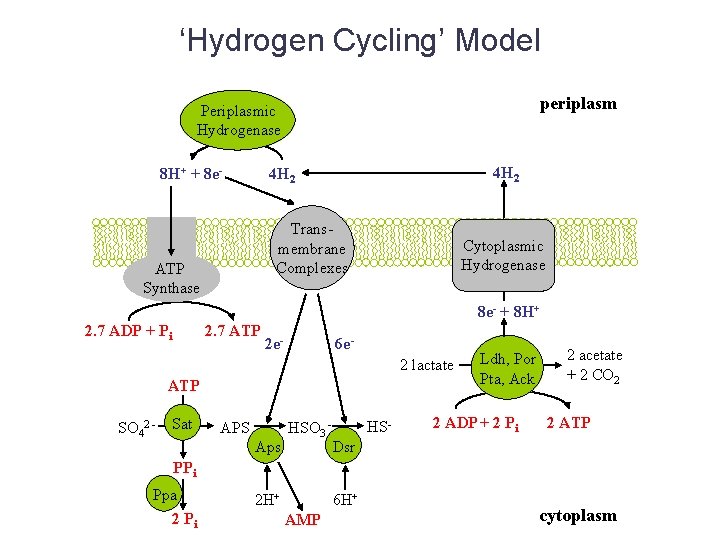
‘Hydrogen Cycling’ Model periplasm Periplasmic Hydrogenase 8 H+ + 8 e- 4 H 2 Transmembrane Complexes ATP Synthase Cytoplasmic Hydrogenase 8 e- + 8 H+ 2. 7 ADP + Pi 2. 7 ATP 2 e- 6 e 2 lactate ATP SO 42 - Sat APS Aps HSO 3 - HS- Ldh, Por Pta, Ack 2 ADP + 2 Pi 2 acetate + 2 CO 2 2 ATP Dsr PPi Ppa 2 Pi 2 H+ 6 H+ AMP cytoplasm
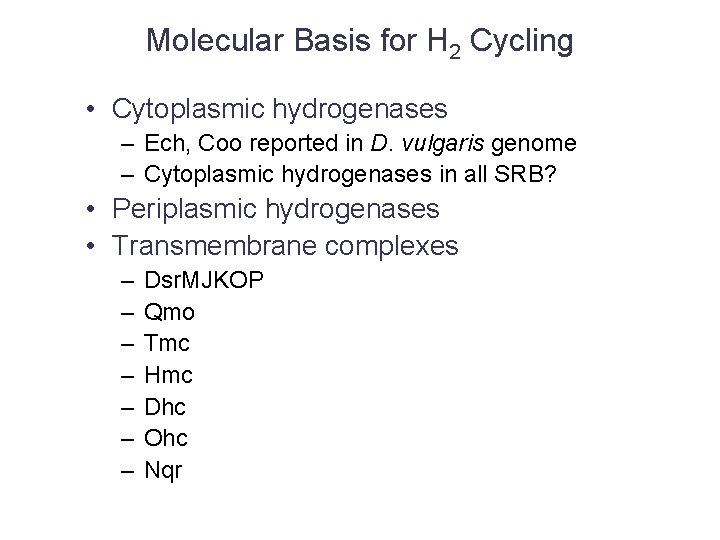
Molecular Basis for H 2 Cycling • Cytoplasmic hydrogenases – Ech, Coo reported in D. vulgaris genome – Cytoplasmic hydrogenases in all SRB? • Periplasmic hydrogenases • Transmembrane complexes – – – – Dsr. MJKOP Qmo Tmc Hmc Dhc Ohc Nqr
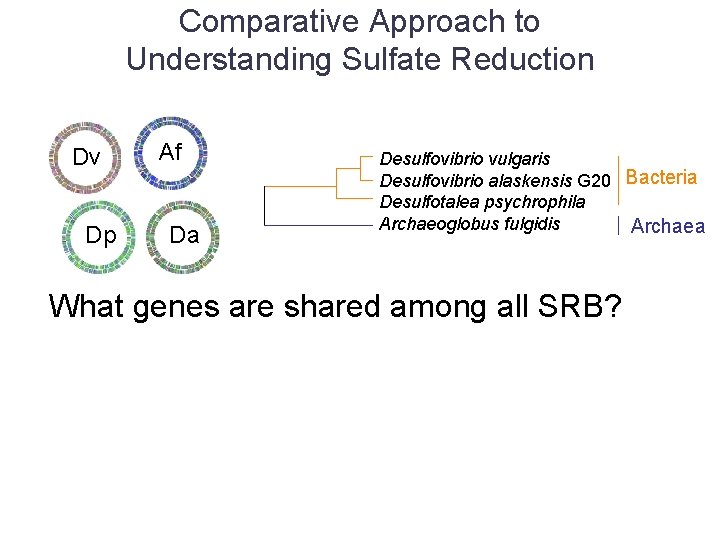
Comparative Approach to Understanding Sulfate Reduction Dv Dp Af Da Desulfovibrio vulgaris Desulfovibrio alaskensis G 20 Bacteria Desulfotalea psychrophila Archaeoglobus fulgidis Archaea What genes are shared among all SRB?
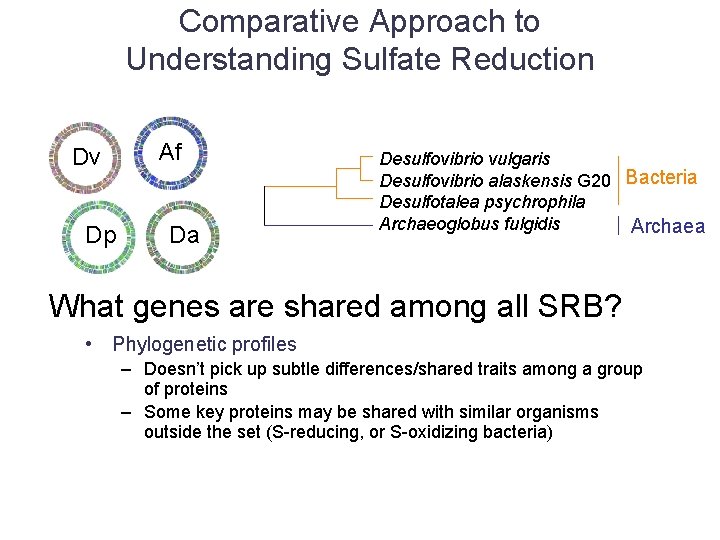
Comparative Approach to Understanding Sulfate Reduction Dv Dp Af Da Desulfovibrio vulgaris Desulfovibrio alaskensis G 20 Bacteria Desulfotalea psychrophila Archaeoglobus fulgidis Archaea What genes are shared among all SRB? • Phylogenetic profiles – Doesn’t pick up subtle differences/shared traits among a group of proteins – Some key proteins may be shared with similar organisms outside the set (S-reducing, or S-oxidizing bacteria)

Comparative Approach to Understanding Sulfate Reduction Dv Dp Af Da Desulfovibrio vulgaris Desulfovibrio alaskensis G 20 Bacteria Desulfotalea psychrophila Archaeoglobus fulgidis Archaea What genes are shared among all SRB? • Phylogenetic profiles – Doesn’t pick up subtle differences/shared traits among a group of proteins – Some key proteins may be shared with similar organisms outside the set (S-reducing, or S-oxidizing bacteria) • Simple homology-based approach – Find genes for which orthologs in the chosen set of bacteria are within top (ten) BLASTp hits for all 200+ genomes

Results: ‘Signature’ SRB Genes Hydrogenases TM complexes Qmo. ABC Dsr. MKJOP (Dahl, et al. , 2005) (Haveman et al. , 2004) Tmc Hmc Decaheme cyt Nqr Octaheme cyt Signature genes Other genes (vulgaris) [Ni. Fe. Se] periplasmic [Fe] cytoplasmic [Ni. Fe] SO 42 - reduction Dsr. AB Dsr. C Aps. AB Ppa. C Sat
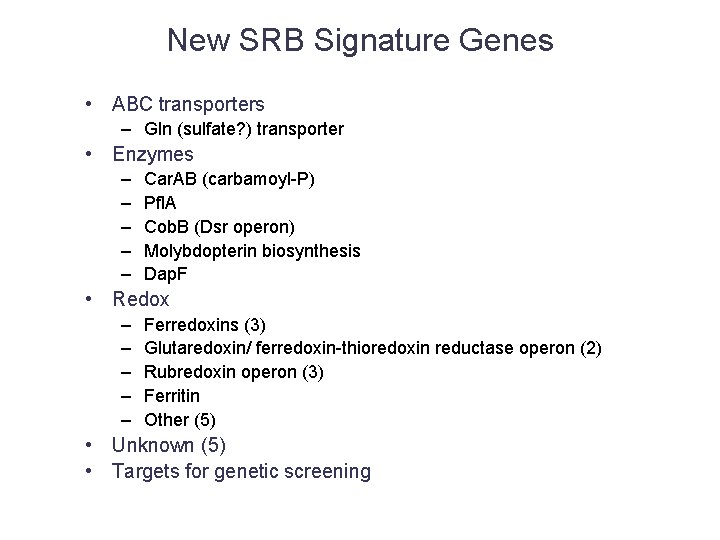
New SRB Signature Genes • ABC transporters – Gln (sulfate? ) transporter • Enzymes – – – Car. AB (carbamoyl-P) Pfl. A Cob. B (Dsr operon) Molybdopterin biosynthesis Dap. F • Redox – – – Ferredoxins (3) Glutaredoxin/ ferredoxin-thioredoxin reductase operon (2) Rubredoxin operon (3) Ferritin Other (5) • Unknown (5) • Targets for genetic screening
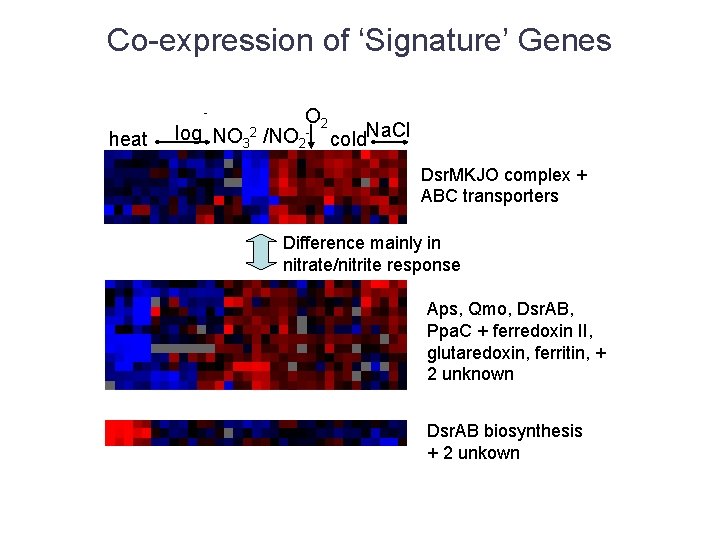
Co-expression of ‘Signature’ Genes - heat O 2 log NO 32 /NO 2 - cold. Na. Cl Dsr. MKJO complex + ABC transporters Difference mainly in nitrate/nitrite response Aps, Qmo, Dsr. AB, Ppa. C + ferredoxin II, glutaredoxin, ferritin, + 2 unknown Dsr. AB biosynthesis + 2 unkown

What’s Not (Exactly) Conserved • H 2 ases – Cytoplasmic • • Dv - Coo, Ech, (fdh) Da - fdh Dp - F 420 -reducing Af - F 420 -reducing – Periplasmic • Various of [Fe], [Ni. Fe. Se] • TM complexes – – – – Qmo (all four species and SOB) Dsr. MKJOP (all four species and SOB) Hmc (Dv, Da) Tmc (Dv, Da) Dhc (Dv, Da) Ohc (Dv) Stc (Dv, Da, Dp)
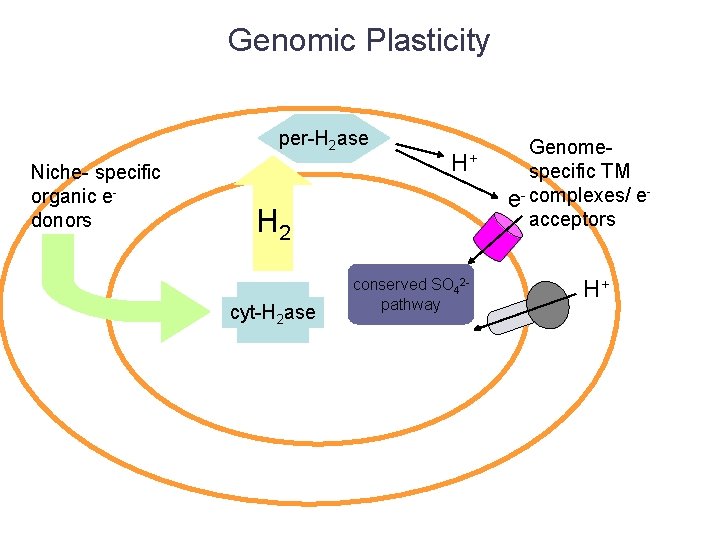
Genomic Plasticity per-H 2 ase Niche- specific organic edonors H+ H 2 cyt-H 2 ase conserved SO 42 pathway Genomespecific TM e- complexes/ eacceptors H+
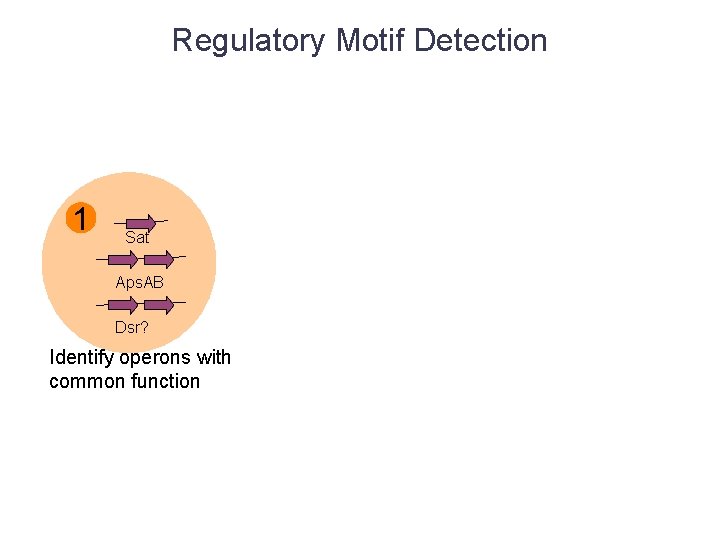
Regulatory Motif Detection 1 Sat Aps. AB Dsr? Identify operons with common function
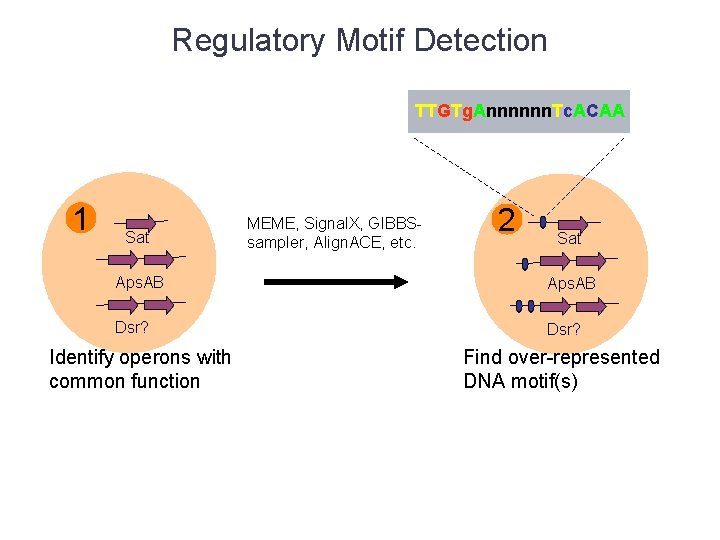
Regulatory Motif Detection TTGTg. Annnnnn. Tc. ACAA 1 Sat MEME, Signal. X, GIBBSsampler, Align. ACE, etc. 2 Sat Aps. AB Dsr? Identify operons with common function Find over-represented DNA motif(s)
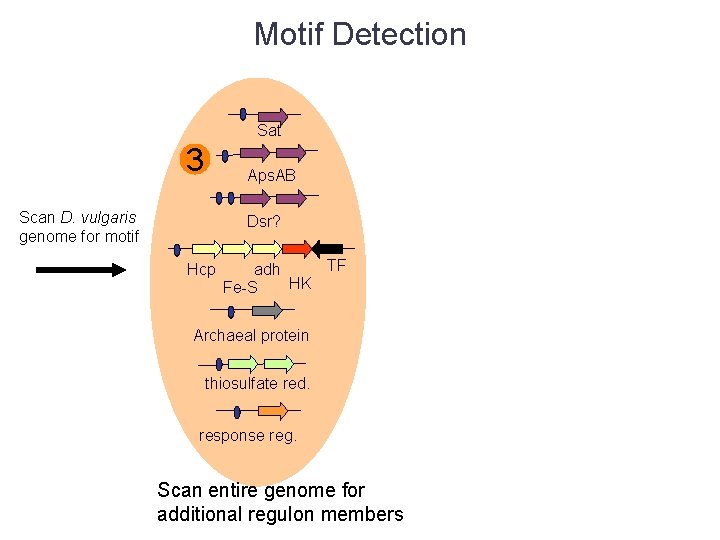
Motif Detection Sat 3 Aps. AB Scan D. vulgaris genome for motif Dsr? Hcp TF adh HK Fe-S Archaeal protein thiosulfate red. response reg. Scan entire genome for additional regulon members
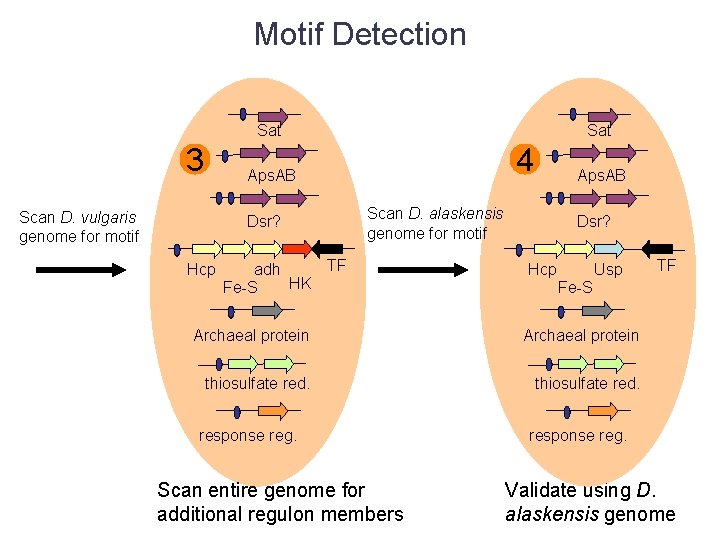
Motif Detection Sat 3 Sat 4 Aps. AB Scan D. vulgaris genome for motif Dsr? Hcp Aps. AB Scan D. alaskensis genome for motif TF adh HK Fe-S Dsr? Hcp Usp Fe-S Archaeal protein thiosulfate red. response reg. Scan entire genome for additional regulon members TF response reg. Validate using D. alaskensis genome
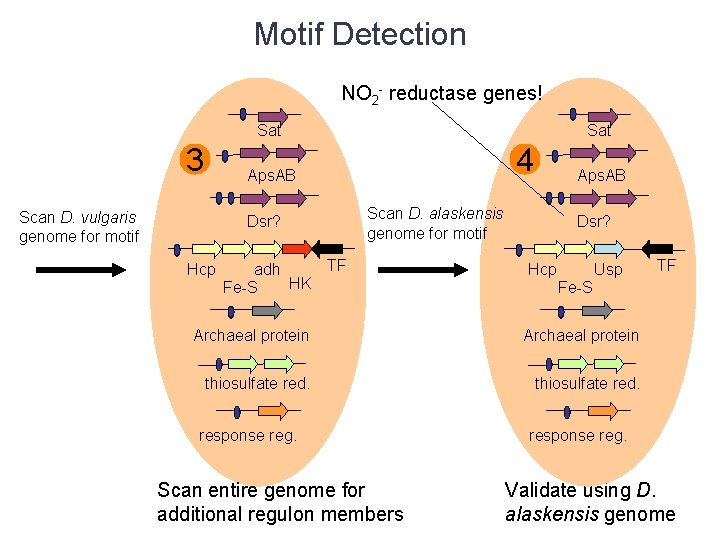
Motif Detection NO 2 - reductase genes! Sat 3 Sat 4 Aps. AB Scan D. vulgaris genome for motif Dsr? Hcp Aps. AB Scan D. alaskensis genome for motif TF adh HK Fe-S Dsr? Hcp Usp Fe-S Archaeal protein thiosulfate red. response reg. Scan entire genome for additional regulon members TF response reg. Validate using D. alaskensis genome

Nitrite Relevant Regulators Hcp. R TTGTg. Annnnnn. Tc. ACAA FUR w. TGAAAatnat. TTTCAw Per. R CAGTAAnnn. TTACTG
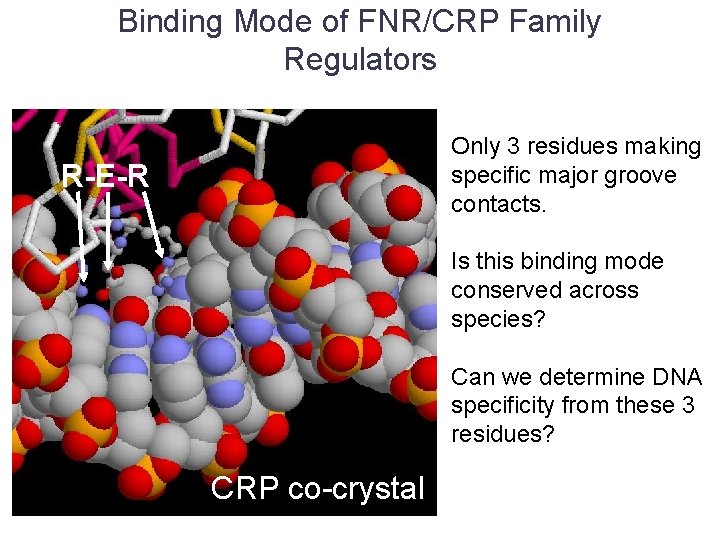
Binding Mode of FNR/CRP Family Regulators Only 3 residues making specific major groove contacts. R-E-R Is this binding mode conserved across species? Can we determine DNA specificity from these 3 residues? CRP co-crystal

CRP/FNR Family Regulators
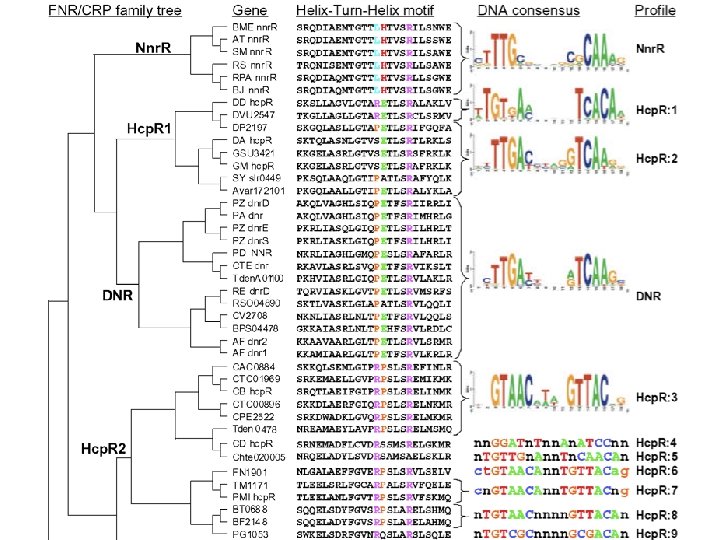

2. 5 m. M Nitrite Response - 60 min Hcp. R Fur
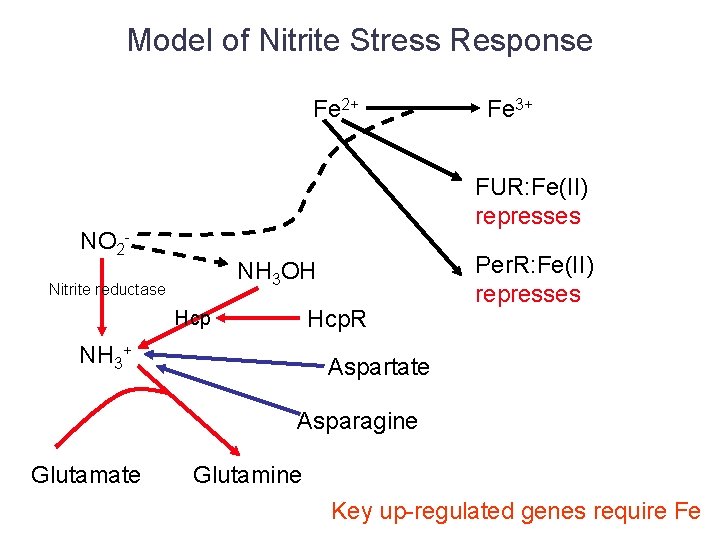
Model of Nitrite Stress Response Fe 2+ Fe 3+ FUR: Fe(II) represses NO 2 - NH 3 OH Nitrite reductase Hcp. R Hcp NH 3+ Per. R: Fe(II) represses Aspartate Asparagine Glutamate Glutamine Key up-regulated genes require Fe

Nitrite Fe Requirement
Experimental Stress Response (O 2) (Rodionov et al. , Genome Biology, 2004) Role for cyd. AB in ‘strict anaerobes’ (Baughn and Malamy, Nature, 2004)
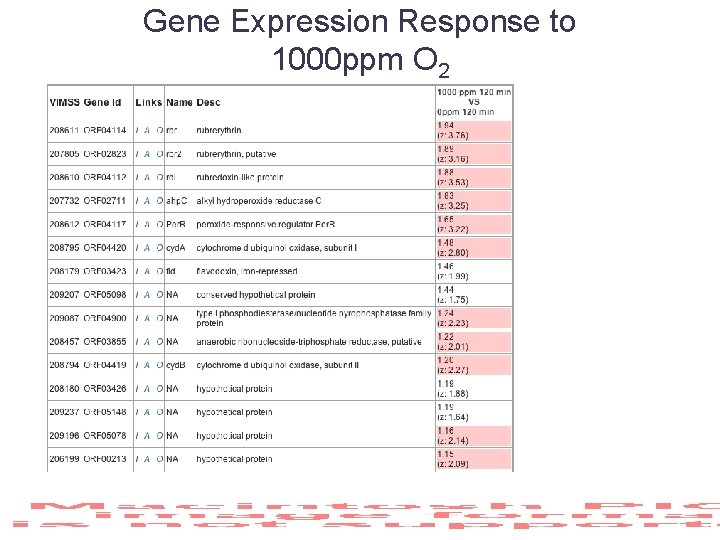
Gene Expression Response to 1000 ppm O 2

Gene Expression Response to 1000 ppm O 2 Cyt-bd oxidase
Environmental Sensing in SRB
Evolution of HPK Signaling Proteins Identify HPK domain proteins 200+ genomes
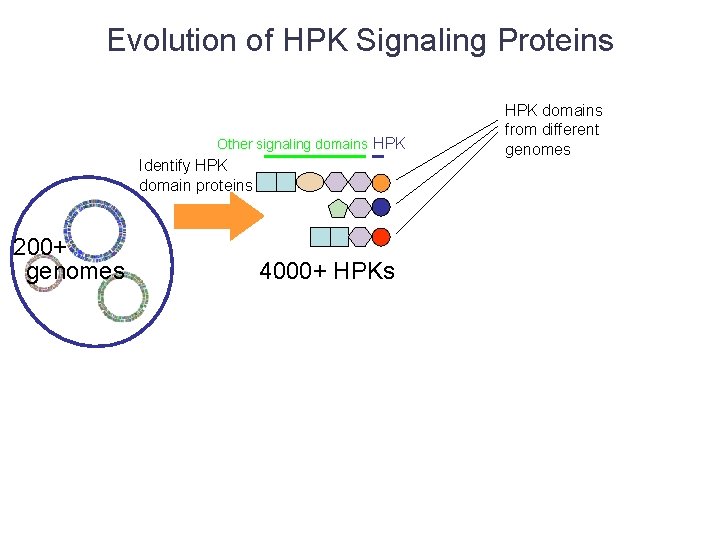
Evolution of HPK Signaling Proteins Other signaling domains HPK Identify HPK domain proteins 200+ genomes 4000+ HPKs HPK domains from different genomes
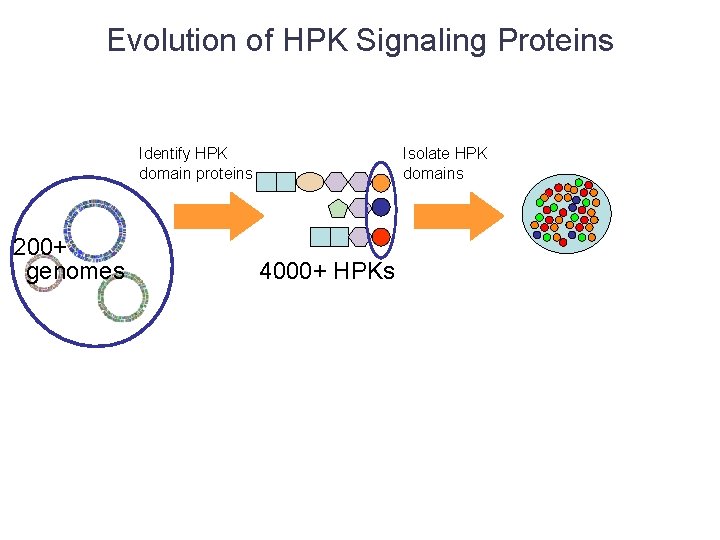
Evolution of HPK Signaling Proteins Identify HPK domain proteins 200+ genomes Isolate HPK domains 4000+ HPKs
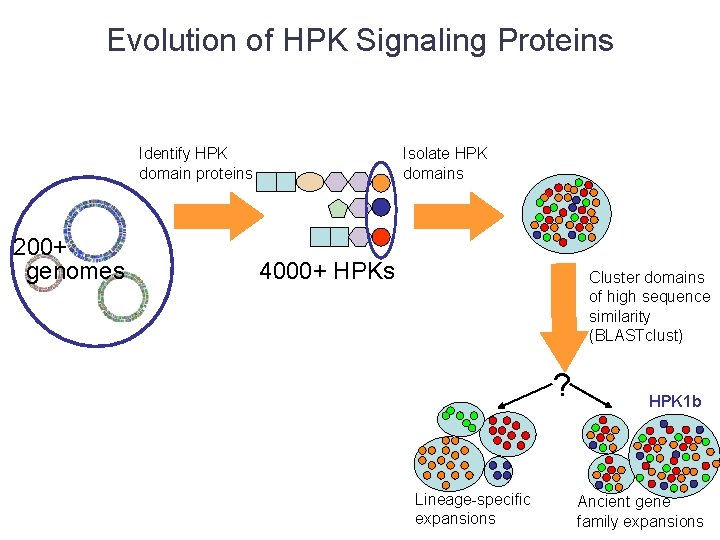
Evolution of HPK Signaling Proteins Identify HPK domain proteins 200+ genomes Isolate HPK domains 4000+ HPKs Cluster domains of high sequence similarity (BLASTclust) ? Lineage-specific expansions HPK 1 b Ancient gene family expansions

NJ tree of HPK 1 b, Cluster 1 of 213 HPKs Caulobacter Gamma D. Vulgaris Vibrio Bdellovibrio Dechloromonas expansion

Cluster 23 H Box TM HAMP PAS HPK che. Y Pseudomonas (closest outgroup) Dv Desulfovibrio expansion HPK G 20
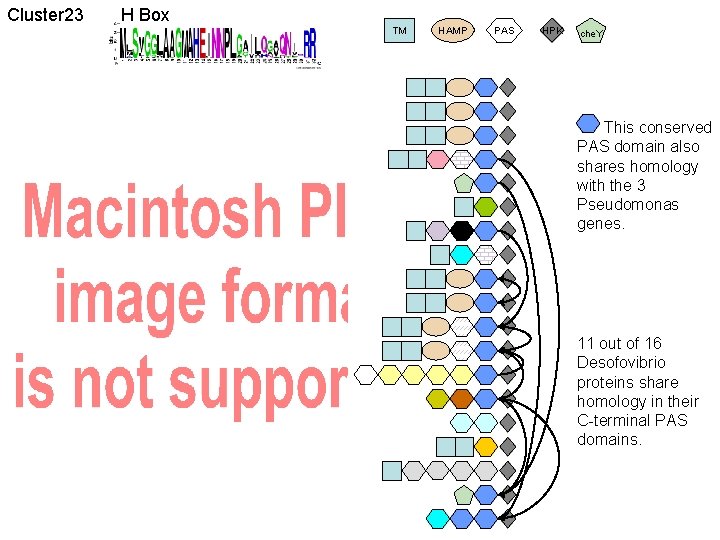
Cluster 23 H Box TM HAMP PAS HPK che. Y This conserved PAS domain also shares homology with the 3 Pseudomonas genes. 11 out of 16 Desofovibrio proteins share homology in their C-terminal PAS domains.
Operons and Bacterial Genomes Functional inference from genomic context New algorithms for functional genomics Bioinformatics methods (clustering) Genome structure and evolution

Operon Evolution • What is the origin of most operons? – HGT (of pre-existing operon) – Novel operon • How/why do novel operons form? – HGT (Selfish Operons) • Do novel operons include HGT genes? • Can ancient (non-HGT) genes form new operons? – Selection for co-regulation • Do operons tend to have more complex regulation?
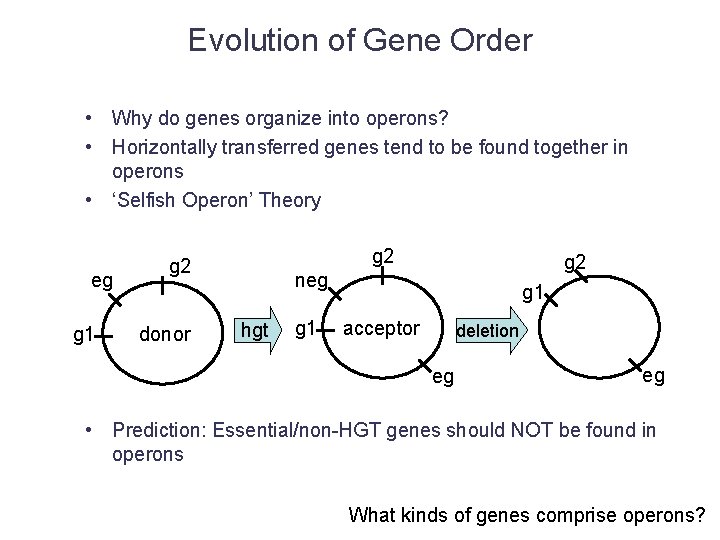
Evolution of Gene Order • Why do genes organize into operons? • Horizontally transferred genes tend to be found together in operons • ‘Selfish Operon’ Theory eg g 1 g 2 donor g 2 neg hgt g 1 acceptor deletion eg eg • Prediction: Essential/non-HGT genes should NOT be found in operons What kinds of genes comprise operons?
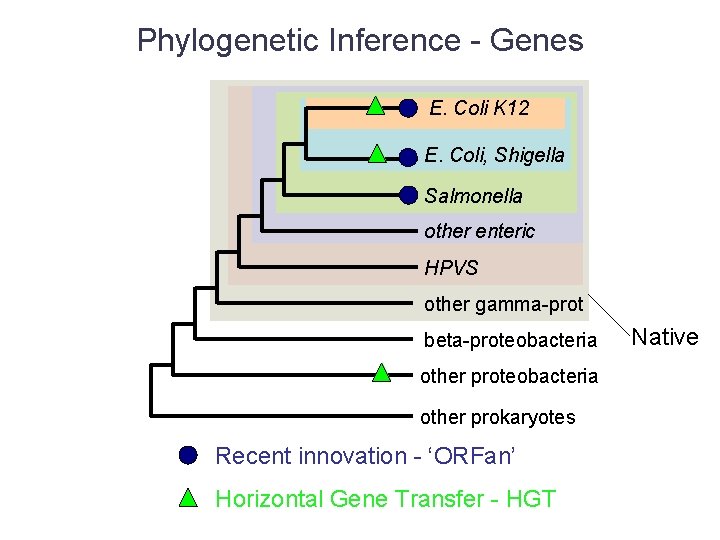
Phylogenetic Inference - Genes E. Coli K 12 E. Coli, Shigella Salmonella other enteric HPVS other gamma-prot beta-proteobacteria other prokaryotes Recent innovation - ‘ORFan’ Horizontal Gene Transfer - HGT Native
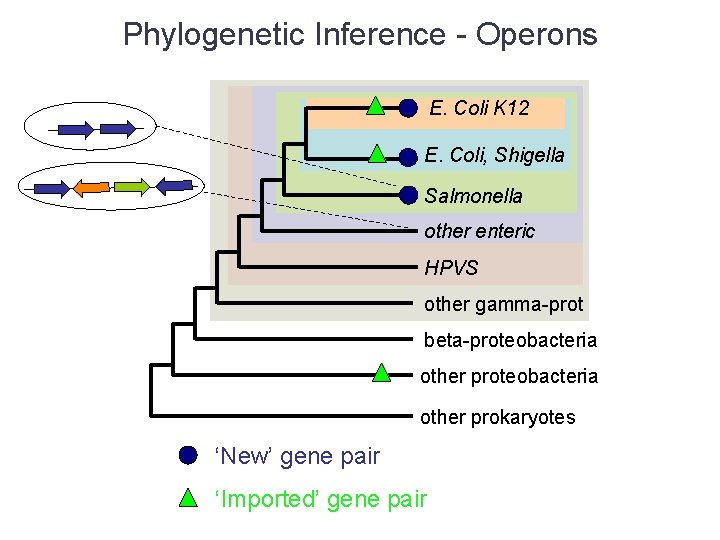
Phylogenetic Inference - Operons E. Coli K 12 E. Coli, Shigella Salmonella other enteric HPVS other gamma-prot beta-proteobacteria other prokaryotes ‘New’ gene pair ‘Imported’ gene pair

No Evidence for Excess HGT in New Operons
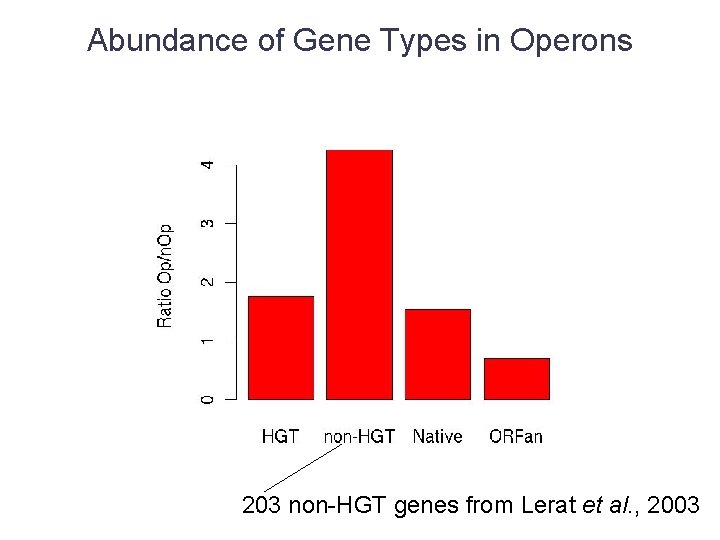
Abundance of Gene Types in Operons 203 non-HGT genes from Lerat et al. , 2003
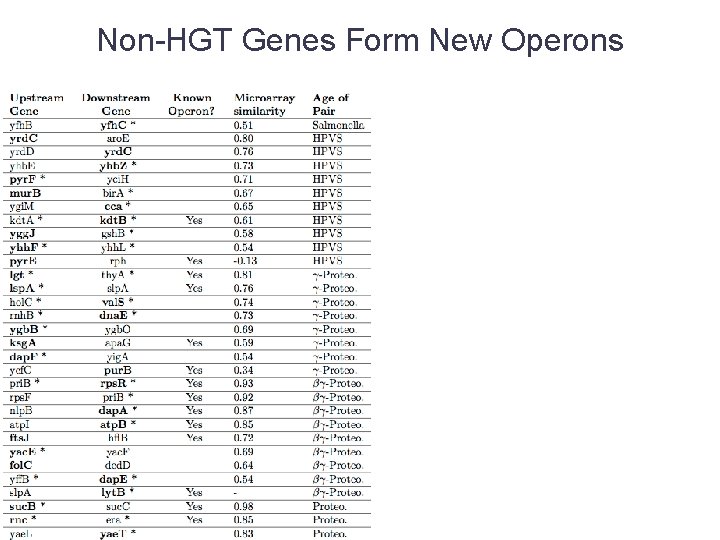
Non-HGT Genes Form New Operons
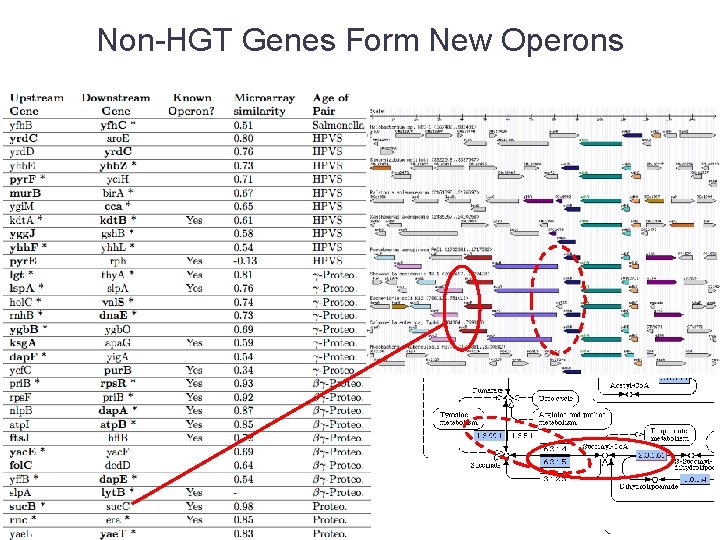
Non-HGT Genes Form New Operons
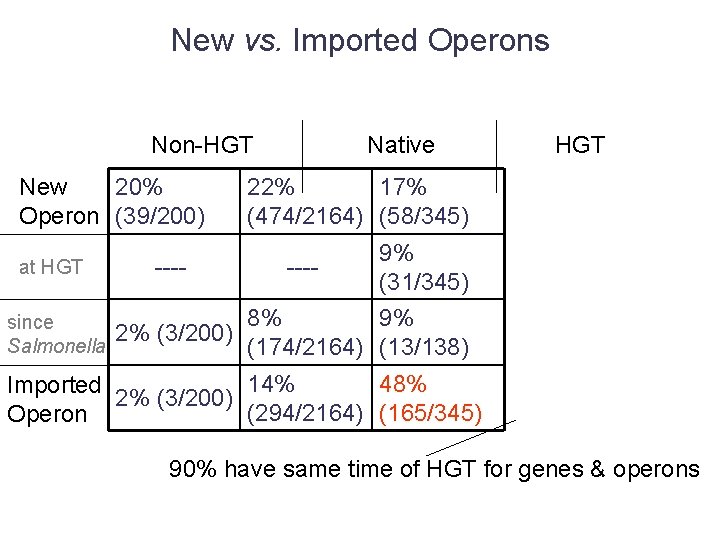
New vs. Imported Operons Non-HGT New 20% Operon (39/200) at HGT since Salmonella ---- Native HGT 22% 17% (474/2164) (58/345) ---- 9% (31/345) 8% 9% 2% (3/200) (174/2164) (13/138) 14% 48% Imported 2% (3/200) (294/2164) (165/345) Operon 90% have same time of HGT for genes & operons
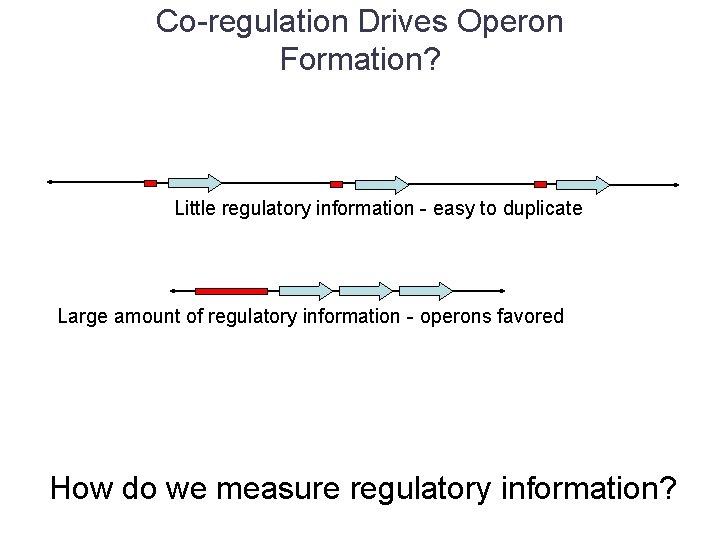
Co-regulation Drives Operon Formation? Little regulatory information - easy to duplicate Large amount of regulatory information - operons favored How do we measure regulatory information?
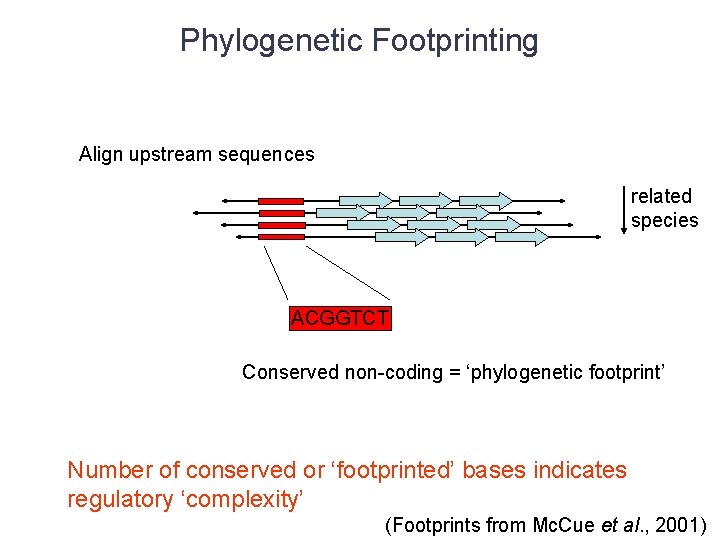
Phylogenetic Footprinting Align upstream sequences related species ACGGTCT Conserved non-coding = ‘phylogenetic footprint’ Number of conserved or ‘footprinted’ bases indicates regulatory ‘complexity’ (Footprints from Mc. Cue et al. , 2001)
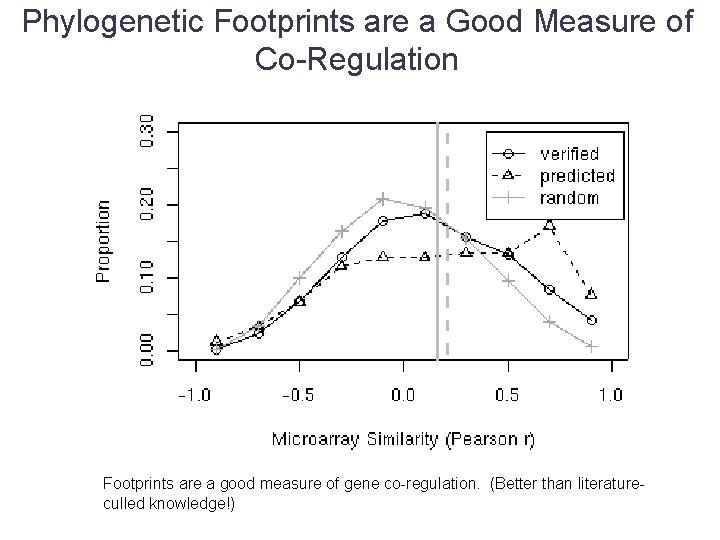
Phylogenetic Footprints are a Good Measure of Co-Regulation Footprints are a good measure of gene co-regulation. (Better than literatureculled knowledge!)
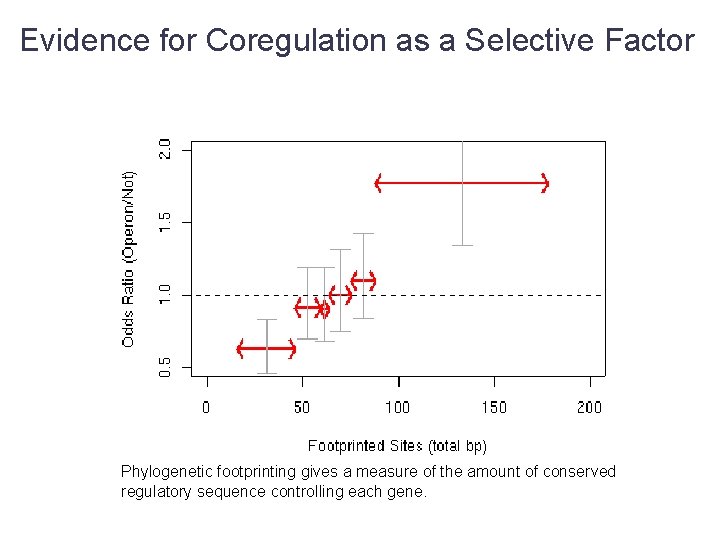
Evidence for Coregulation as a Selective Factor Phylogenetic footprinting gives a measure of the amount of conserved regulatory sequence controlling each gene.

Acknowledgments • Adam Arkin • Computational Core – Katherine Huang – Morgan Price – Keith Keller • Inna Dubchak • Mikhail Gelfand • Dmitry Rodionov • VIMSS – – – – Terry Hazen Lab Jay Keasling Lab Anup Singh Lab Aindrila Mukhopadhyay Judy Wall David Stahl Sergey Stolyar • Genome Annotation – – – Kennan Kellaris Ralf Rabus Gerrit Voordouw Shelley Haveman Larry Barton
- Slides: 54