Subsampled brain MRI reconstruction by generative adversarial neural

Subsampled brain MRI reconstruction by generative adversarial neural networks Roy Shaul, Itamar David, Ohad Shitrit, Tammy Riklin Raviv Tutor: Dr. Shahrooz Faghih Roohi Student: Yuxiang Zheng Data: 14. Jan. 2021
Introduction of MRI A strong magnetic field and pulses of radio frequency (RF) electromagnetic radiation is generated Hydrogen atoms, which are prevalent in living organisms, emit absorbed RF energy “Magnetic Resonance Imaging” Source: https: //commons. wikimedia. org/wiki/File: Siemens_Magnetom_Ae ra_MRI_scanner. jpg#/media/File: Siemens_Magnetom_Aera_MRI_scann er. jpg RF energy is received by antennas Computer Aided Medical Procedures December 13, 2021 Slide 2
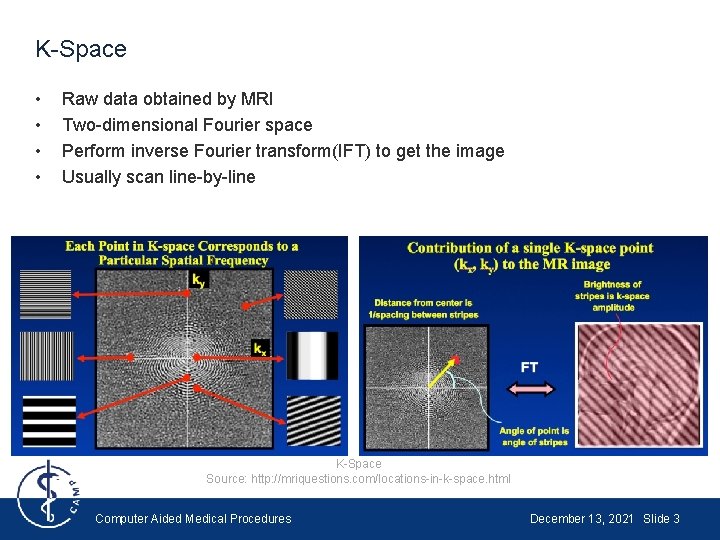
K-Space • • Raw data obtained by MRI Two-dimensional Fourier space Perform inverse Fourier transform(IFT) to get the image Usually scan line-by-line K-Space Source: http: //mriquestions. com/locations-in-k-space. html Computer Aided Medical Procedures December 13, 2021 Slide 3
The challenges of MRI: Acceleration • Long scanning process reduces patient comfort • For cases where motion cannot be reduced, motion artifacts will occur • For dynamic scanning, time resolution and spatial resolution need to be balanced Computer Aided Medical Procedures December 13, 2021 Slide 4
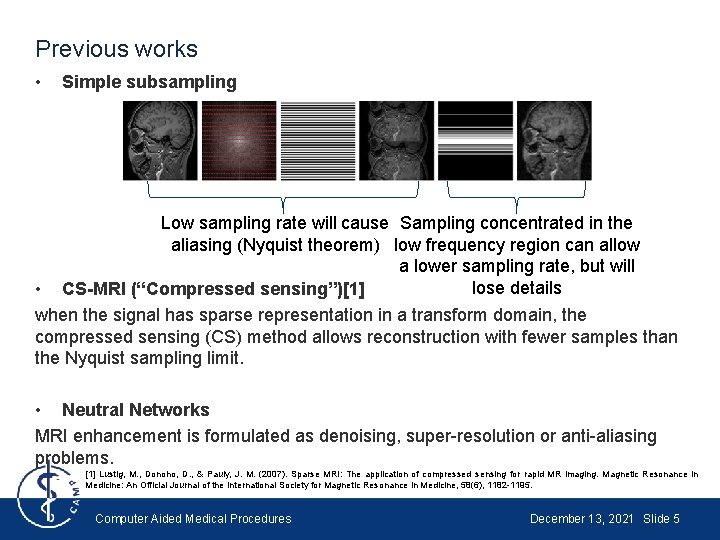
Previous works • Simple subsampling Low sampling rate will cause Sampling concentrated in the aliasing (Nyquist theorem) low frequency region can allow a lower sampling rate, but will lose details CS-MRI (“Compressed sensing”)[1] • when the signal has sparse representation in a transform domain, the compressed sensing (CS) method allows reconstruction with fewer samples than the Nyquist sampling limit. • Neutral Networks MRI enhancement is formulated as denoising, super-resolution or anti-aliasing problems. [1] Lustig, M. , Donoho, D. , & Pauly, J. M. (2007). Sparse MRI: The application of compressed sensing for rapid MR imaging. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine, 58(6), 1182 -1195. Computer Aided Medical Procedures December 13, 2021 Slide 5
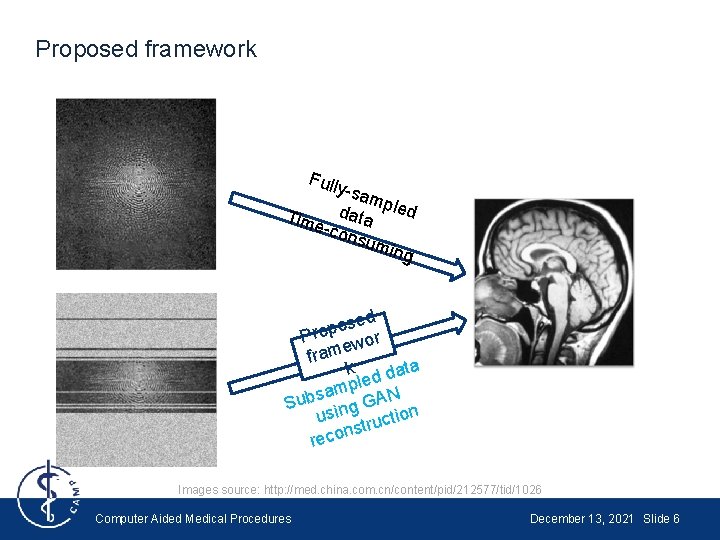
Proposed framework Full y-sa mpl ed Tim data e-co nsu min g osed p o r P wor e m fra k ata d d e mpl N a s b GA n Su g n i us ctio u r t s n reco Images source: http: //med. china. com. cn/content/pid/212577/tid/1026 Computer Aided Medical Procedures December 13, 2021 Slide 6
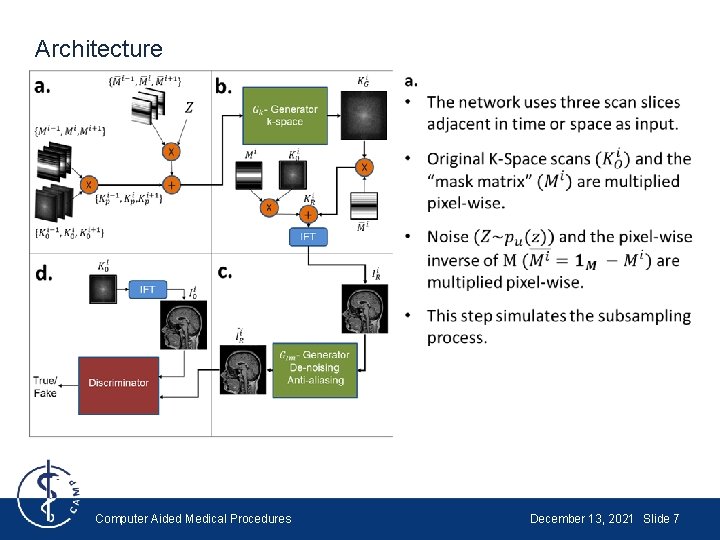
Architecture Computer Aided Medical Procedures December 13, 2021 Slide 7
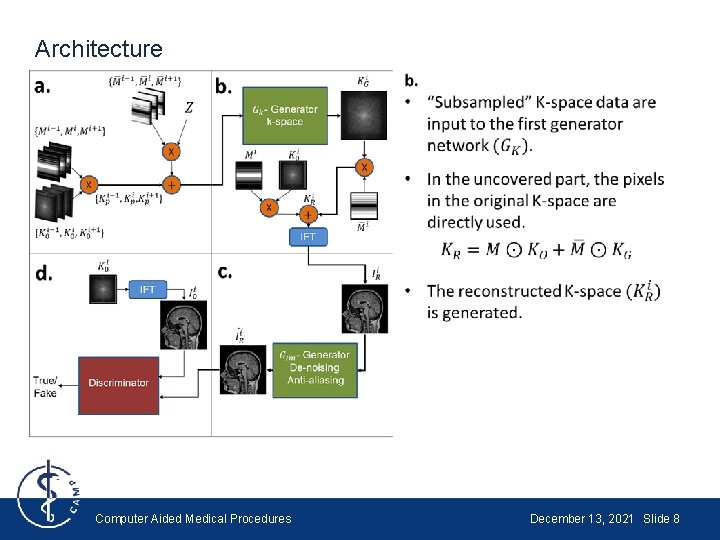
Architecture Computer Aided Medical Procedures December 13, 2021 Slide 8
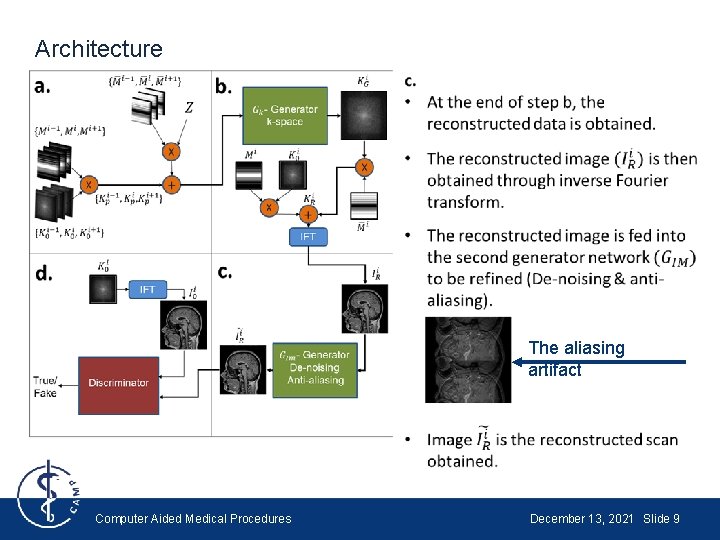
Architecture The aliasing artifact Computer Aided Medical Procedures December 13, 2021 Slide 9
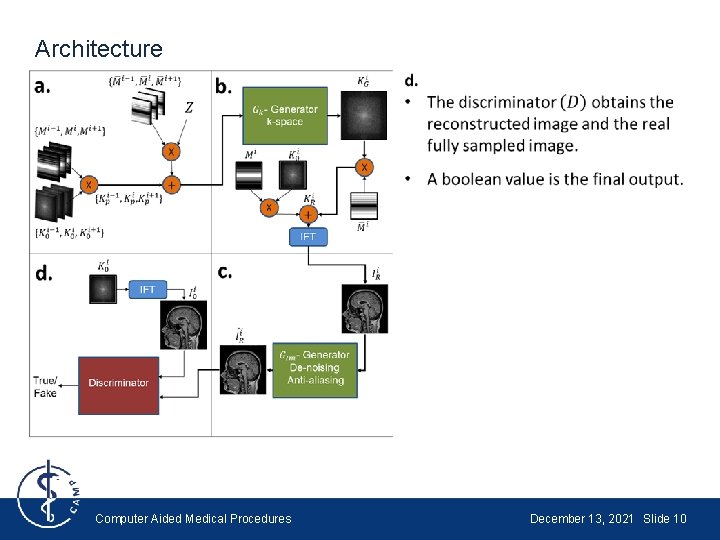
Architecture Computer Aided Medical Procedures December 13, 2021 Slide 10
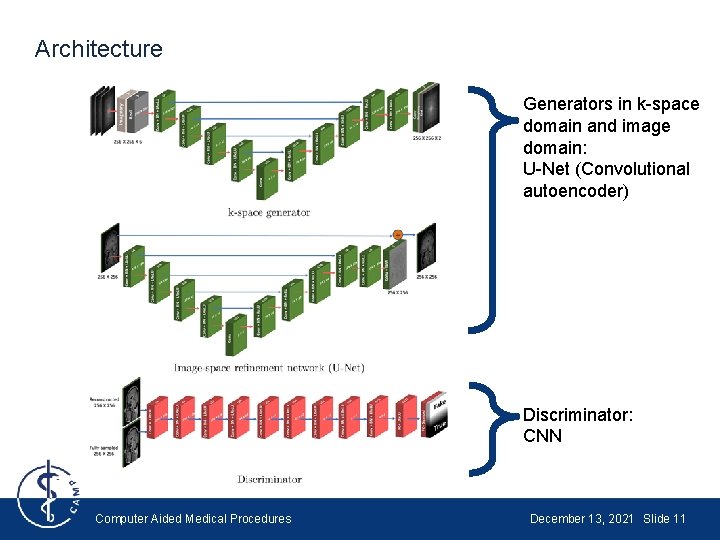
Architecture Generators in k-space domain and image domain: U-Net (Convolutional autoencoder) Discriminator: CNN Computer Aided Medical Procedures December 13, 2021 Slide 11
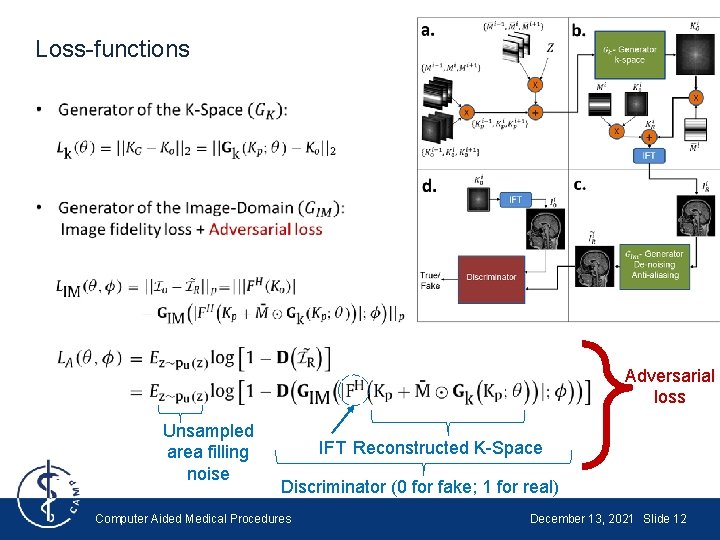
Loss-functions Adversarial loss Unsampled area filling noise IFT Reconstructed K-Space Discriminator (0 for fake; 1 for real) Computer Aided Medical Procedures December 13, 2021 Slide 12
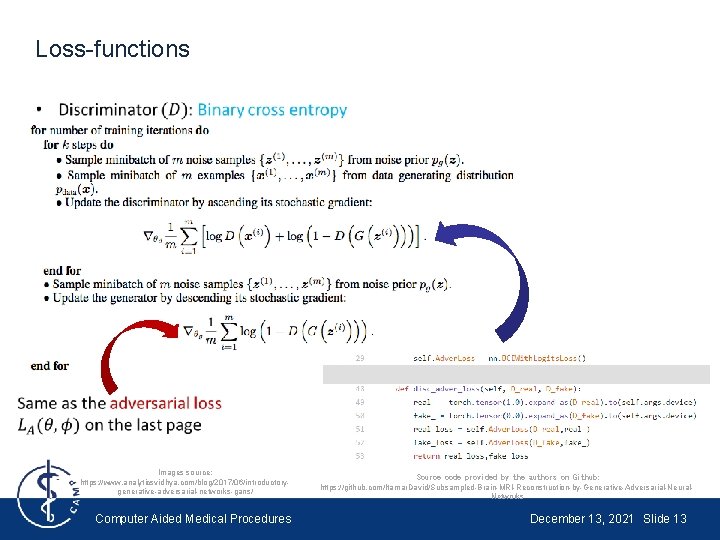
Loss-functions Images source: https: //www. analyticsvidhya. com/blog/2017/06/introductorygenerative-adversarial-networks-gans/ Computer Aided Medical Procedures Source code provided by the authors on Github: https: //github. com/Itamar. David/Subsampled-Brain-MRI-Reconstruction-by-Generative-Adversarial-Neural. Networks December 13, 2021 Slide 13
Training process details • Generators training process is considered: • Reason: At the early stage of training, the discriminator can easily distinguish fake samples, so this part has almost no contribution to the convergence. Therefore, training the generator in advance can effectively accelerate the overall convergence of the network. Computer Aided Medical Procedures December 13, 2021 Slide 14
![Evaluation parameters • [2] Wang, Z. , Bovik, A. C. , Sheikh, H. R. Evaluation parameters • [2] Wang, Z. , Bovik, A. C. , Sheikh, H. R.](http://slidetodoc.com/presentation_image_h2/05e72de38684236a68a7bcb42f0627b4/image-15.jpg)
Evaluation parameters • [2] Wang, Z. , Bovik, A. C. , Sheikh, H. R. , & Simoncelli, E. P. (2004). Image quality assessment: from error visibility to structural similarity. IEEE transactions on image processing, 13(4), 600 -612. [3] Dubuisson, M. P. , & Jain, A. K. (1994, October). A modified Hausdorff distance for object matching. In Proceedings of 12 th international conference on pattern recognition (Vol. 1, pp. 566 -568). IEEE. Computer Aided Medical Procedures December 13, 2021 Slide 15
![Comparison researches • [4] Yang, Y. , Sun, J. , Li, H. , & Comparison researches • [4] Yang, Y. , Sun, J. , Li, H. , &](http://slidetodoc.com/presentation_image_h2/05e72de38684236a68a7bcb42f0627b4/image-16.jpg)
Comparison researches • [4] Yang, Y. , Sun, J. , Li, H. , & Xu, Z. (2018). ADMM-CSNet: A deep learning approach for image compressive sensing. IEEE transactions on pattern analysis and machine intelligence. Computer Aided Medical Procedures December 13, 2021 Slide 16

Datasets • • • IXI Dataset: Brain scans of healthy adults 2015 Longitudinal MS Lesion Segmentation Challenge [5]: Brain scans of multiple sclerosis patients is included, using four sequences (T 1 -w, T 2 -w, PDw and FLAIR) DCE-MRI Dataset: Brain scans of stroke and brain tumor patients Optimizer • Adams [5] Carass, A. , Roy, S. , Jog, A. , Cuzzocreo, J. L. , Magrath, E. , Gherman, A. , . . . & Crainiceanu, C. M. (2017). Longitudinal multiple sclerosis lesion segmentation data resource. Data in brief, 12, 346 -350. Computer Aided Medical Procedures December 13, 2021 Slide 17
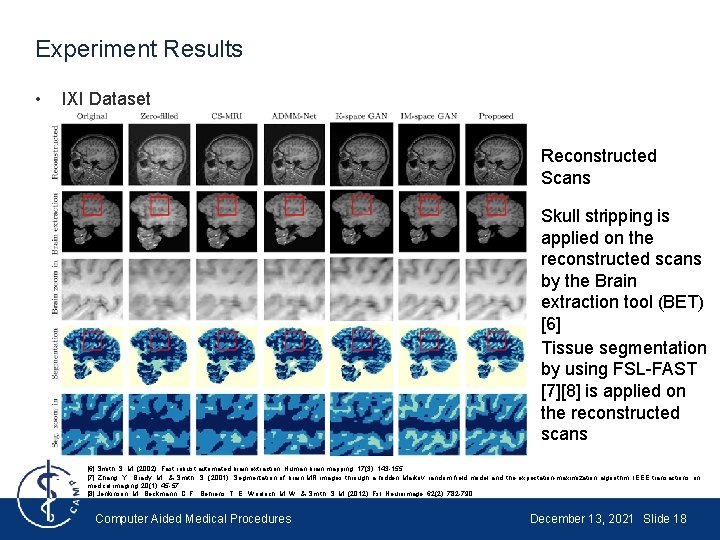
Experiment Results • IXI Dataset Reconstructed Scans Skull stripping is applied on the reconstructed scans by the Brain extraction tool (BET) [6] Tissue segmentation by using FSL-FAST [7][8] is applied on the reconstructed scans [6] Smith, S. M. (2002). Fast robust automated brain extraction. Human brain mapping, 17(3), 143 -155. [7] Zhang, Y. , Brady, M. , & Smith, S. (2001). Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE transactions on medical imaging, 20(1), 45 -57. [8] Jenkinson, M. , Beckmann, C. F. , Behrens, T. E. , Woolrich, M. W. , & Smith, S. M. (2012). Fsl. Neuroimage, 62(2), 782 -790. Computer Aided Medical Procedures December 13, 2021 Slide 18
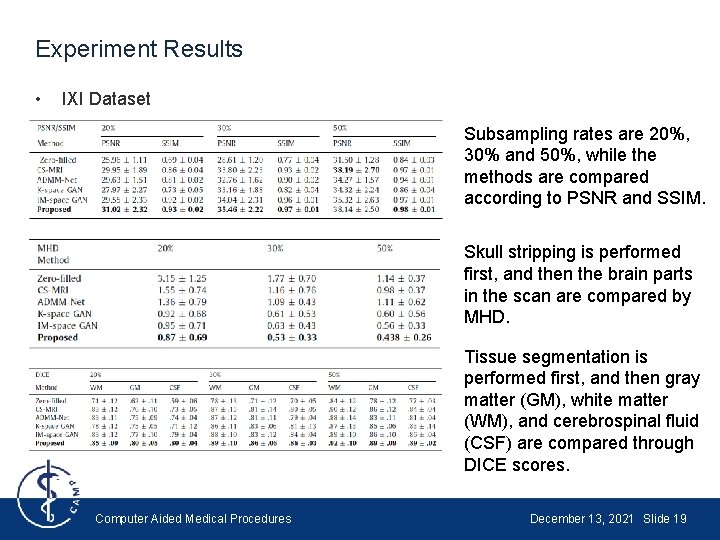
Experiment Results • IXI Dataset Subsampling rates are 20%, 30% and 50%, while the methods are compared according to PSNR and SSIM. Skull stripping is performed first, and then the brain parts in the scan are compared by MHD. Tissue segmentation is performed first, and then gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF) are compared through DICE scores. Computer Aided Medical Procedures December 13, 2021 Slide 19
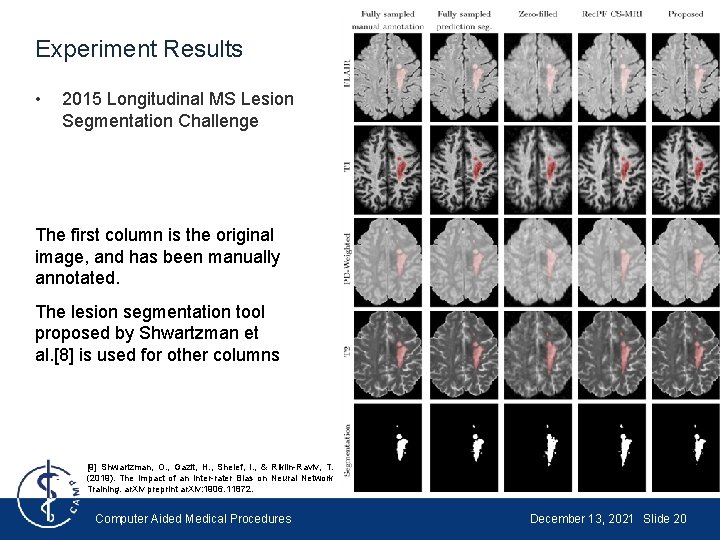
Experiment Results • 2015 Longitudinal MS Lesion Segmentation Challenge The first column is the original image, and has been manually annotated. The lesion segmentation tool proposed by Shwartzman et al. [8] is used for other columns [9] Shwartzman, O. , Gazit, H. , Shelef, I. , & Riklin-Raviv, T. (2019). The Impact of an Inter-rater Bias on Neural Network Training. ar. Xiv preprint ar. Xiv: 1906. 11872. Computer Aided Medical Procedures December 13, 2021 Slide 20
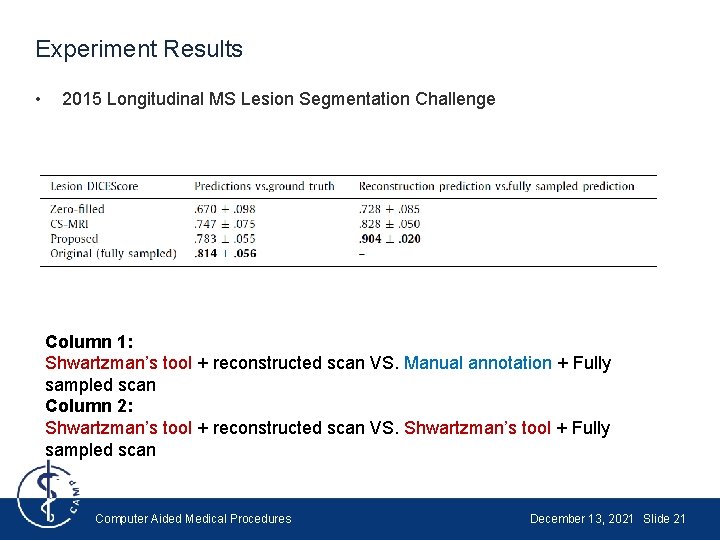
Experiment Results • 2015 Longitudinal MS Lesion Segmentation Challenge Column 1: Shwartzman’s tool + reconstructed scan VS. Manual annotation + Fully sampled scan Column 2: Shwartzman’s tool + reconstructed scan VS. Shwartzman’s tool + Fully sampled scan Computer Aided Medical Procedures December 13, 2021 Slide 21
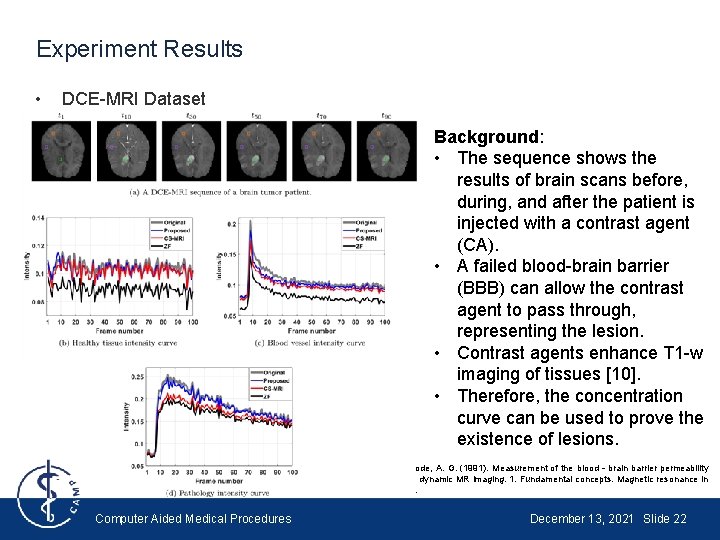
Experiment Results • DCE-MRI Dataset Background: • The sequence shows the results of brain scans before, during, and after the patient is injected with a contrast agent (CA). • A failed blood-brain barrier (BBB) can allow the contrast agent to pass through, representing the lesion. • Contrast agents enhance T 1 -w imaging of tissues [10]. • Therefore, the concentration curve can be used to prove the existence of lesions. [10] Tofts, P. S. , & Kermode, A. G. (1991). Measurement of the blood‐brain barrier permeability and leakage space using dynamic MR imaging. 1. Fundamental concepts. Magnetic resonance in medicine, 17(2), 357 -367. Computer Aided Medical Procedures December 13, 2021 Slide 22
![Experiment Results • DCE-MRI Dataset [11] Tofts, P. S. (1997). Modeling tracer kinetics in Experiment Results • DCE-MRI Dataset [11] Tofts, P. S. (1997). Modeling tracer kinetics in](http://slidetodoc.com/presentation_image_h2/05e72de38684236a68a7bcb42f0627b4/image-23.jpg)
Experiment Results • DCE-MRI Dataset [11] Tofts, P. S. (1997). Modeling tracer kinetics in dynamic Gd‐DTPA MR imaging. Journal of magnetic resonance imaging, 7(1), 91 -101. [12] Tofts, P. S. (2010). T 1 -weighted DCE imaging concepts: modelling, acquisition and analysis. signal, 500(450), 400. Computer Aided Medical Procedures December 13, 2021 Slide 23

Conclusion • • • The proposed method is a two-stage GAN framework for estimating the missing k-space samples and refining in the image-space. The network can reconstruct the original scan from highly subsampled kspace data. In the experiment, the minimum proportion of k-space data used is 20%. The authors' experiments are based on a variety of datasets and include multiple modes of MRI scan (T 1 -w, T 2 -w, PD, and FLAIR). The authors claim that the solution is “real-time software-only”. The evaluation parameters used include not only the evaluation of image fidelity, but also the evaluation of clinical usability. Future work • The authors mentioned that they hope to consider clinical usability in the loss function. That is, the clinical usability should be quantified through a certain parameter and directly reflected in the loss function used for network training. Computer Aided Medical Procedures December 13, 2021 Slide 24

Personal overview Contribution: • The authors propose a GAN-based subsampling MRI reconstruction tool, which can greatly speed up the MRI scanning process and does not require additional hardware assistance. • For brain scans, the authors designed detailed and rigorous experiments. Weakness: • The authors did not provide information about training time and hardware resources. • The experimental results given in the article are all for brain scans, and do not show the effect of the network on other body parts. Especially in nonstationary parts such as the heart, or parts with large individual differences. • Even if this method can be used for other parts of the body, it needs to provide corresponding dataset for training. Computer Aided Medical Procedures December 13, 2021 Slide 25
![Another related work Accelerated Magnetic Resonance Imaging by Adversarial Neural Network [13] proposed by Another related work Accelerated Magnetic Resonance Imaging by Adversarial Neural Network [13] proposed by](http://slidetodoc.com/presentation_image_h2/05e72de38684236a68a7bcb42f0627b4/image-26.jpg)
Another related work Accelerated Magnetic Resonance Imaging by Adversarial Neural Network [13] proposed by Ohad Shitrit, Tammy Riklin Raviv (two authors of this paper) in 2017 • Architecture Generator Discriminator • Experiment result (with 52% subsampled K-Space) [13] Shitrit, O. , & Raviv, T. R. (2017). Accelerated magnetic resonance imaging by adversarial neural network. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support (pp. 30 -38). Springer, Cham. Computer Aided Medical Procedures December 13, 2021 Slide 26

Thank you for your attention! Any Questions?
- Slides: 27