Study and engineering of gene function mutagenesis I





- Slides: 38

Study and engineering of gene function: mutagenesis I. II. Why mutagenize? Random mutagenesis, mutant selection schemes III. Site-directed mutagenesis, deletion mutagenesis IV. Engineering of proteins V. Alterations in the genetic code Course Packet: #30

Uses for mutagenesis • Define the role of a gene--are phenotypes altered by mutations? • Determine functionally important regions of a gene (in vivo or in vitro) • Improve or change the function of a gene product • Investigate functions of non-genes, eg. DNA regions important for regulation

Protein engineering-Why? • Enhance stability/function under new conditions – temperature, p. H, organic/aqueous solvent, [salt] • Alter enzyme substrate specificity • Enhance enzymatic rate • Alter epitope binding properties

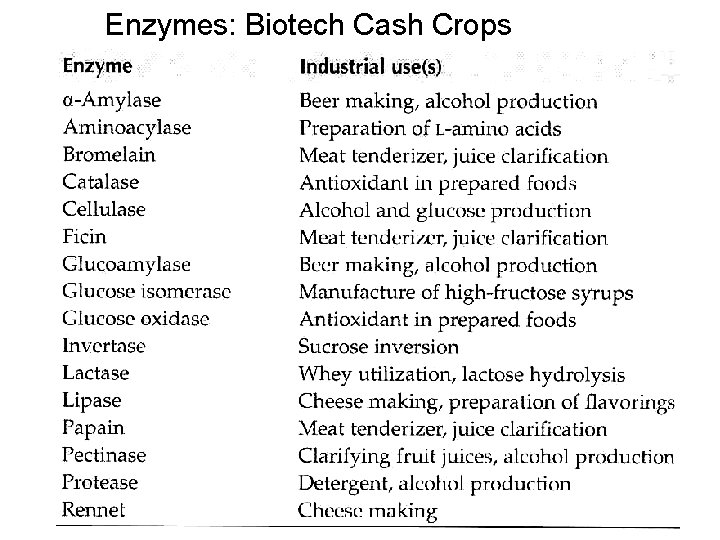
Enzymes: Biotech Cash Crops

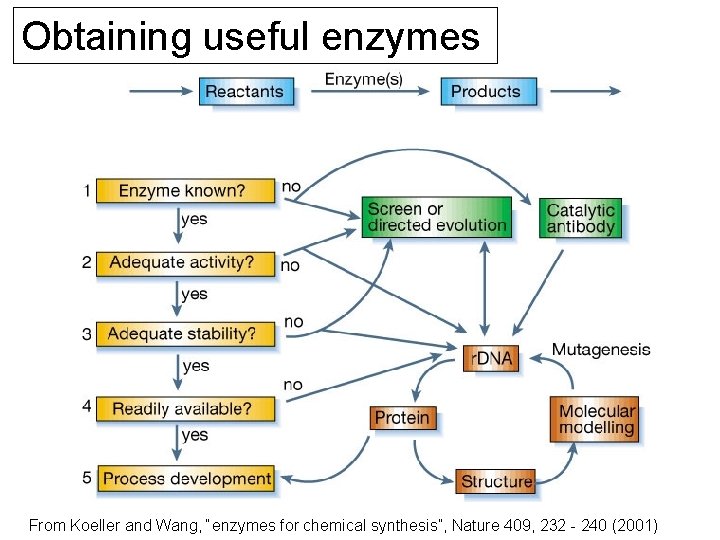
Obtaining useful enzymes From Koeller and Wang, “enzymes for chemical synthesis”, Nature 409, 232 - 240 (2001)

Random mutagenesis • Cassette mutagenesis with “doped”oligos • Chemical mutagenesis – expose short piece of DNA to mutagen, make “library” of clones, test for phenotypes • PCR mutagenesis by base misincorporation – Include Mn 2+ in reaction – Reduce concentration of one d. NTP

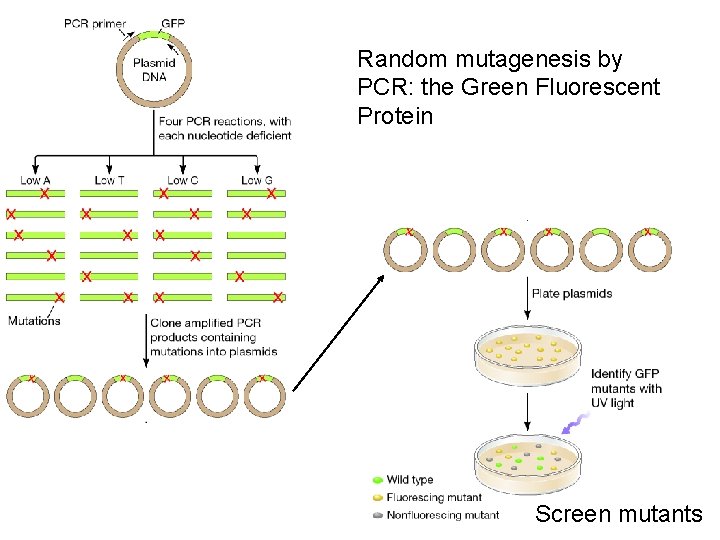
Random mutagenesis by PCR: the Green Fluorescent Protein Screen mutants

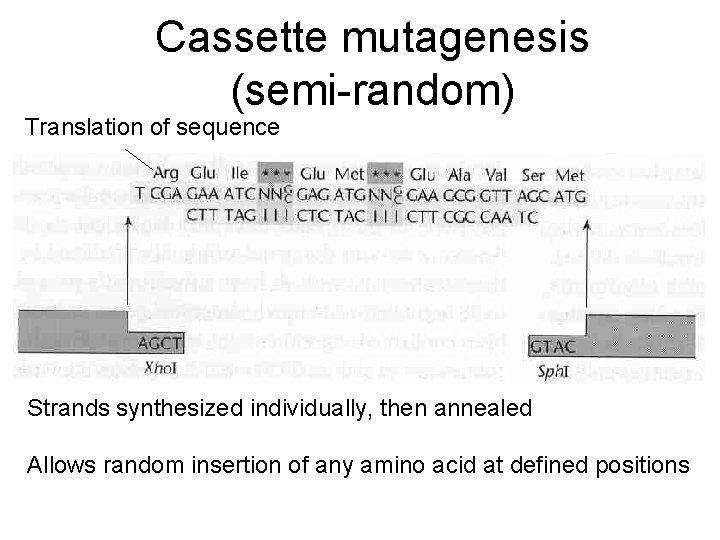
Cassette mutagenesis (semi-random) Translation of sequence Strands synthesized individually, then annealed Allows random insertion of any amino acid at defined positions

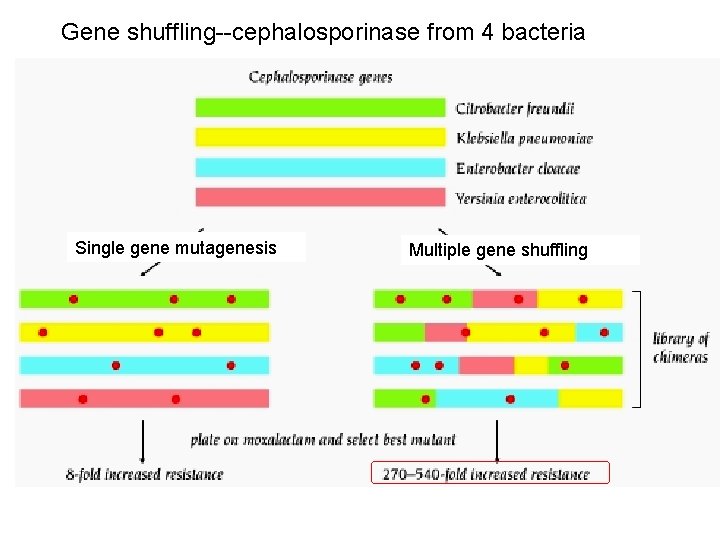
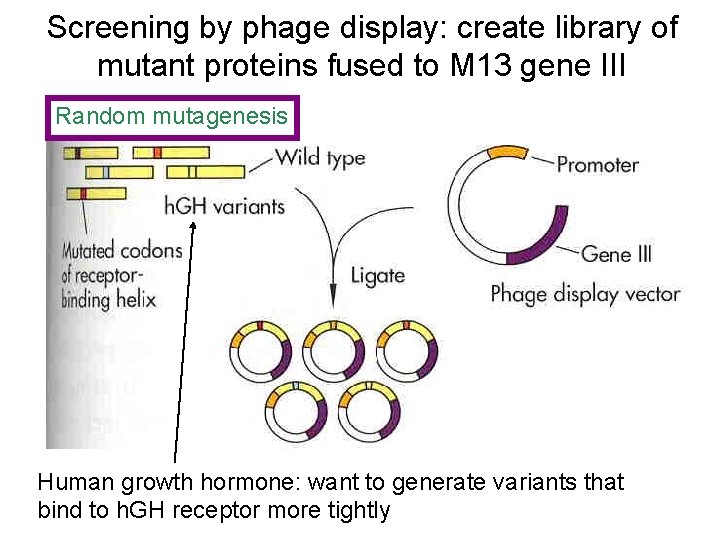
Random and semi-random mutagenesis: directed evolution • Mutagenize existing protein, eg. error-prone PCR, doped oligo cassette mutagenesis -- and/or -Do “gene shuffling” (Creates Library) • Screen library of mutations for proteins with altered properties – Standard screening: 10, 000 - 100, 000 mutants – Phage display: 109 mutants

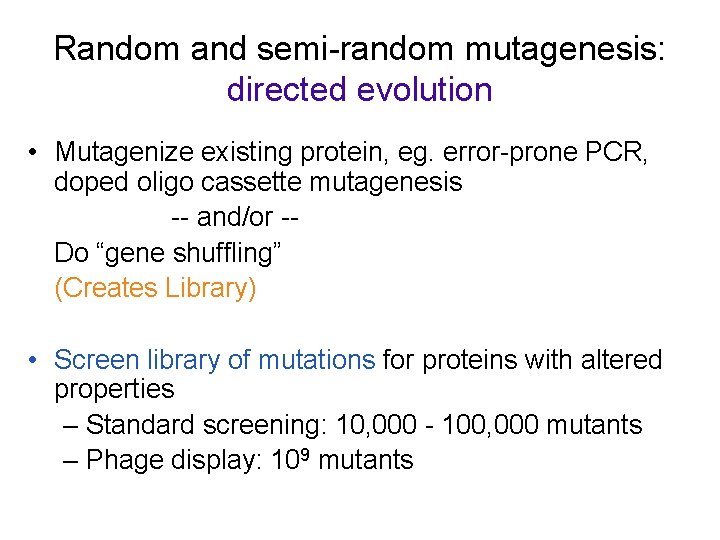
Gene shuffling: “sexual PCR”

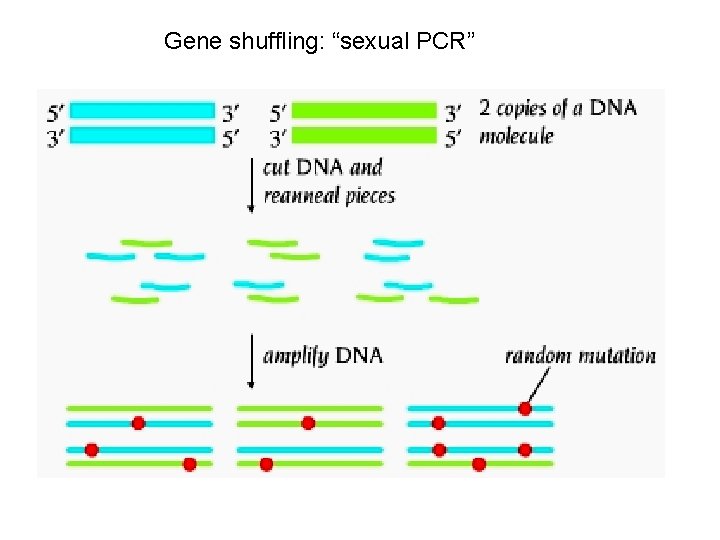
Gene shuffling For gene shuffling protocols you must have related genes in original pool: 1) evolutionary variants, or 2) variants mutated in vitro Shuffling allows rapid scanning through sequence space: faster than doing multiple rounds of random mutagenesis and screening

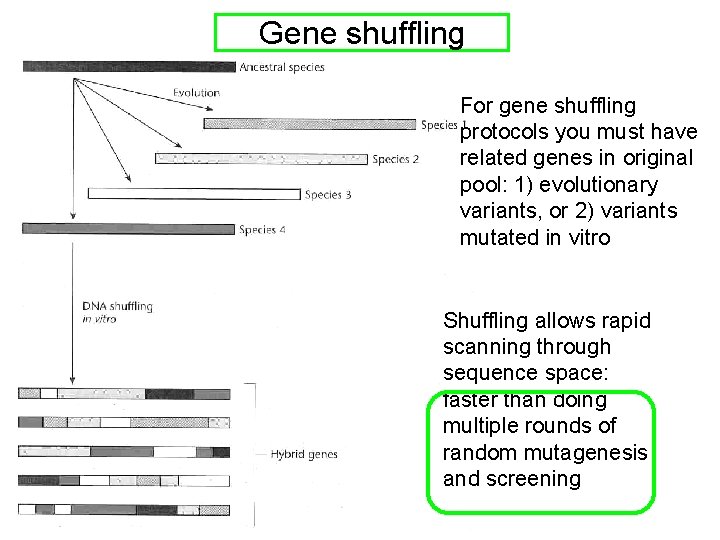
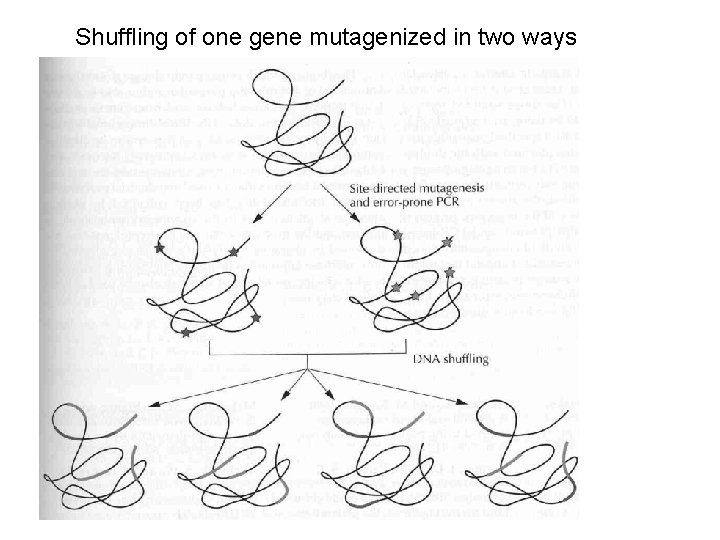
Shuffling of one gene mutagenized in two ways

Gene shuffling--cephalosporinase from 4 bacteria Single gene mutagenesis Multiple gene shuffling

Screening by phage display: create library of mutant proteins fused to M 13 gene III Random mutagenesis Human growth hormone: want to generate variants that bind to h. GH receptor more tightly

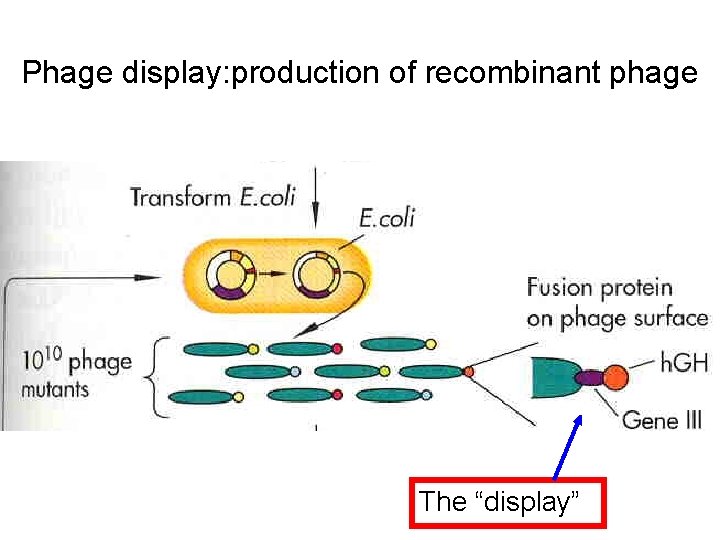
Phage display: production of recombinant phage The “display”

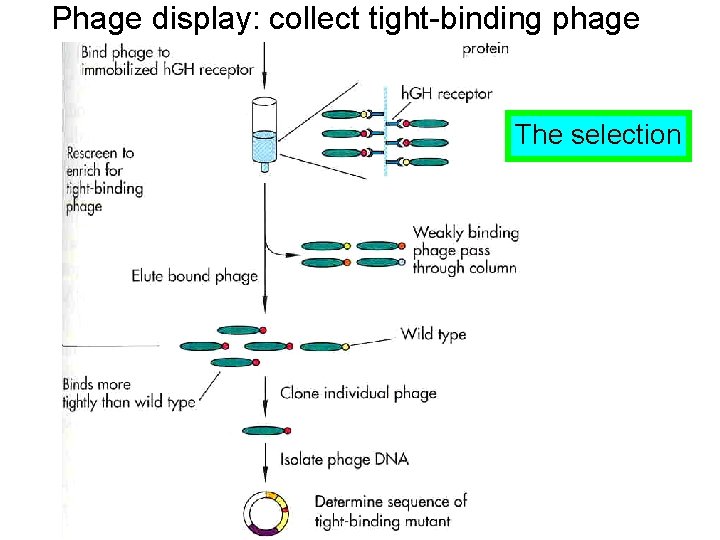
Phage display: collect tight-binding phage The selection

Animation of phage display http: //www. dyax. com/discovery/phagedisplay. html

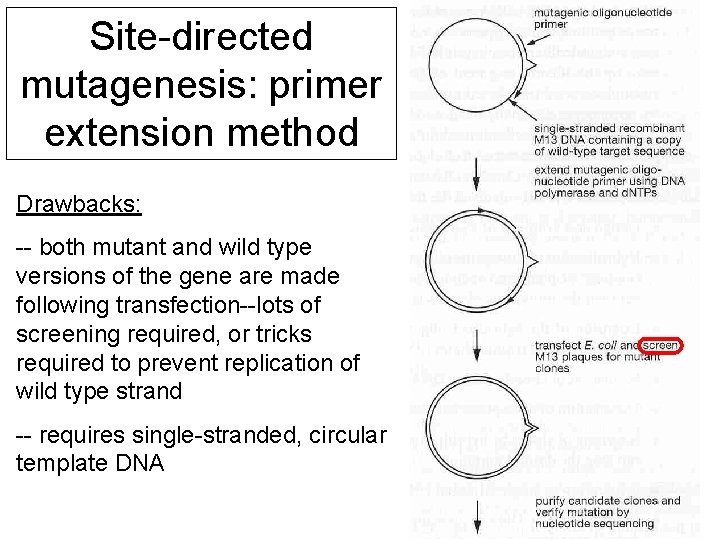
Site-directed mutagenesis: primer extension method Drawbacks: -- both mutant and wild type versions of the gene are made following transfection--lots of screening required, or tricks required to prevent replication of wild type strand -- requires single-stranded, circular template DNA

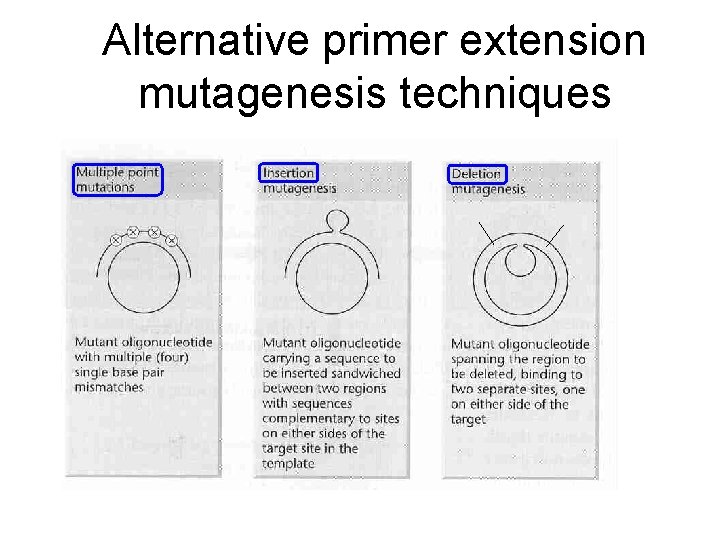
Alternative primer extension mutagenesis techniques

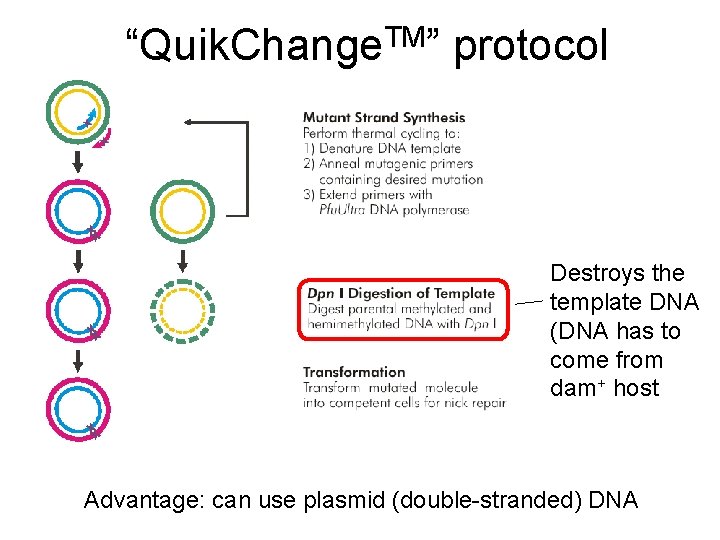
TM “Quik. Change ” protocol Destroys the template DNA (DNA has to come from dam+ host Advantage: can use plasmid (double-stranded) DNA

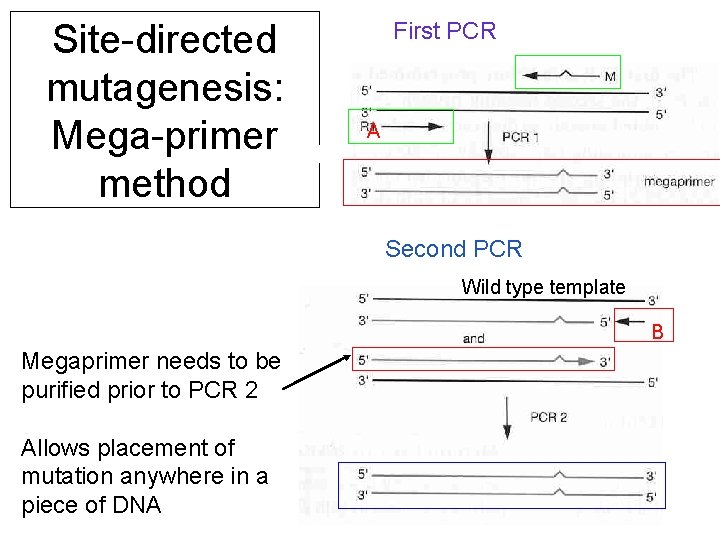
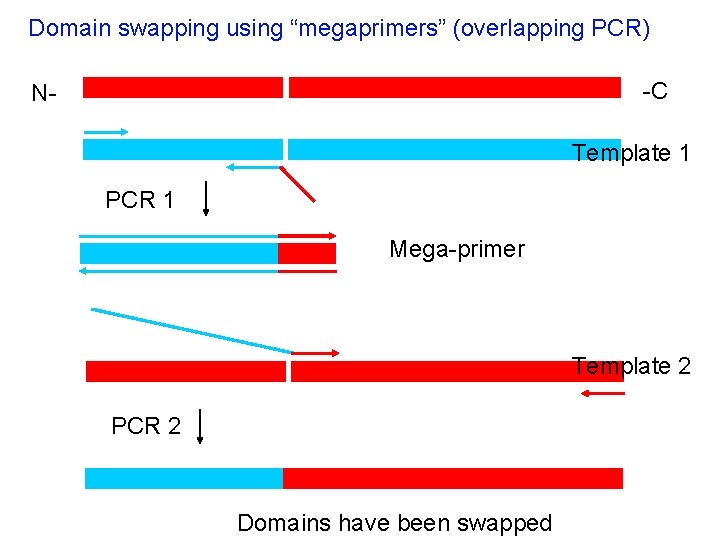
Site-directed mutagenesis: Mega-primer method First PCR A Second PCR Wild type template B Megaprimer needs to be purified prior to PCR 2 Allows placement of mutation anywhere in a piece of DNA

Domain swapping using “megaprimers” (overlapping PCR) -C N- Template 1 PCR 1 Mega-primer Template 2 PCR 2 Domains have been swapped

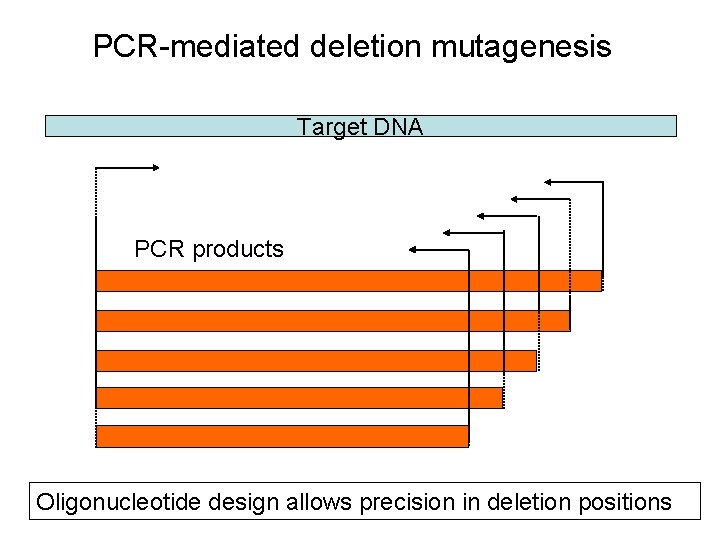
PCR-mediated deletion mutagenesis Target DNA PCR products Oligonucleotide design allows precision in deletion positions

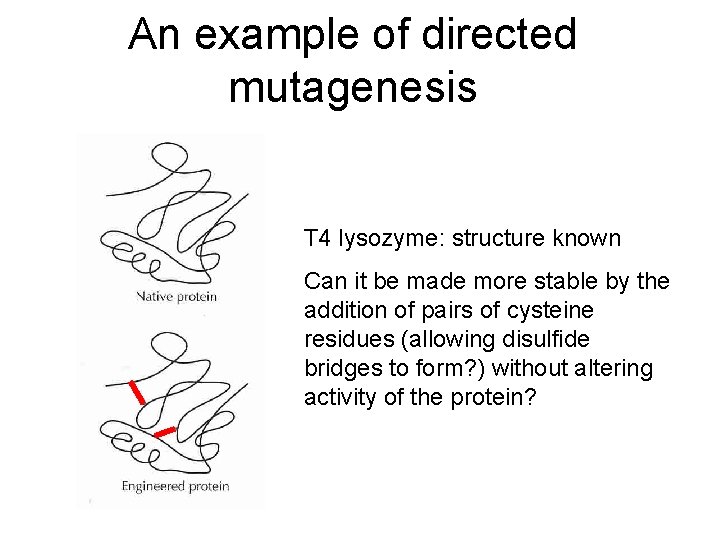
Directed mutagenesis • Make changes in amino acid sequence based on rational decisions • Structure known? Mutate amino acids in any part of protein thought to influence activity/stability/solubility etc. • Protein with multiple family members? Mutate desired protein in positions that bring it closer to another family member with desired properties

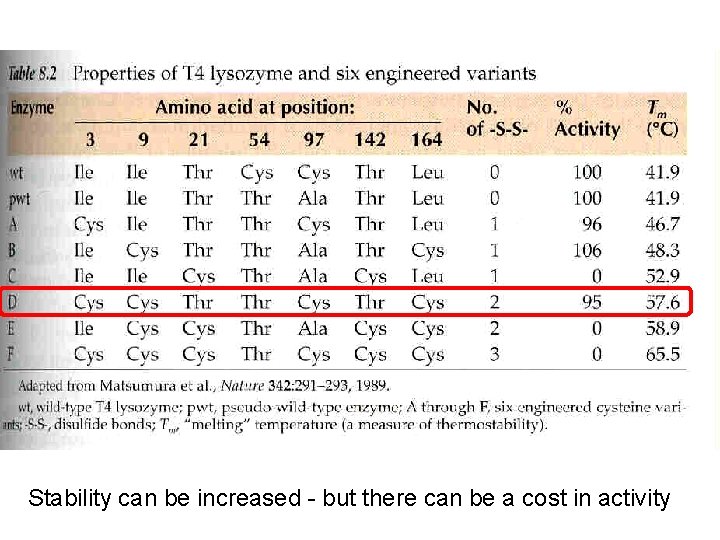
An example of directed mutagenesis T 4 lysozyme: structure known Can it be made more stable by the addition of pairs of cysteine residues (allowing disulfide bridges to form? ) without altering activity of the protein?

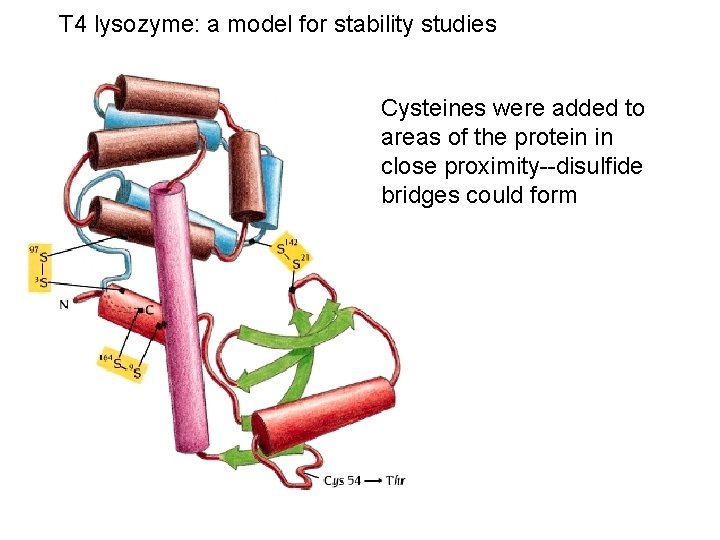
T 4 lysozyme: a model for stability studies Cysteines were added to areas of the protein in close proximity--disulfide bridges could form

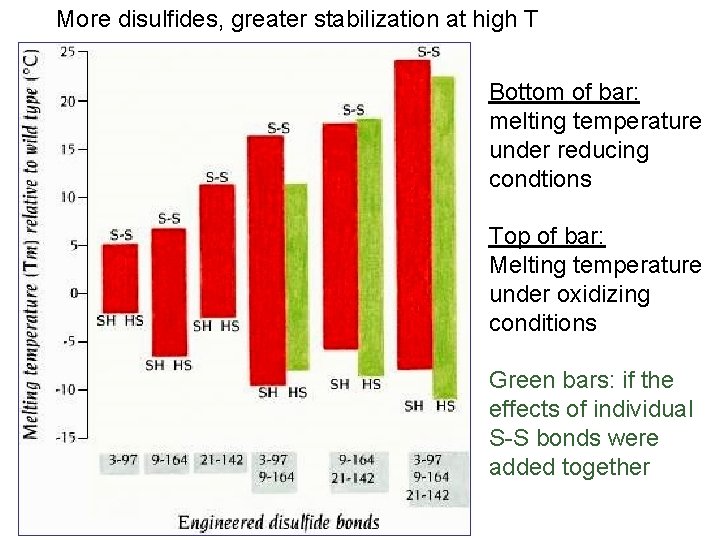
More disulfides, greater stabilization at high T Bottom of bar: melting temperature under reducing condtions Top of bar: Melting temperature under oxidizing conditions Green bars: if the effects of individual S-S bonds were added together

Stability can be increased - but there can be a cost in activity

The genetic code • 61 sense codons, 3 non-sense (stop) codons • 20 amino acids • Other amino acids, some in the cell (as precursors to other amino acids), but very rarely have any been added to the genetic code in a living system • Is it possible to add new amino acids to the code? • Yes. . . sort of Wang et al. (2001) “Expanding the genetic code” Science 292, p. 498.

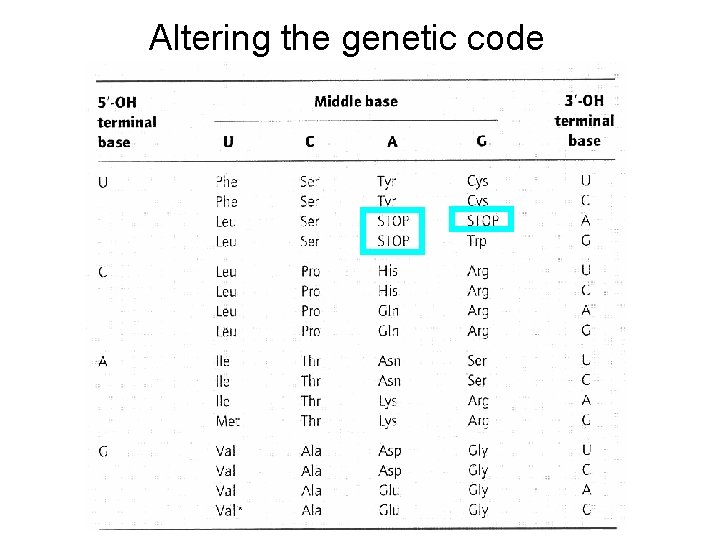
Altering the genetic code

Why add new amino acids to proteins? • New amino acid = new functional group • Alter or enhance protein function (rational design) • Chemically modify protein following synthesis (chemical derivitization) – Probe protein structure, function – Modify protein in vivo, add labels and monitor protein localization, movement, dynamics in living cells

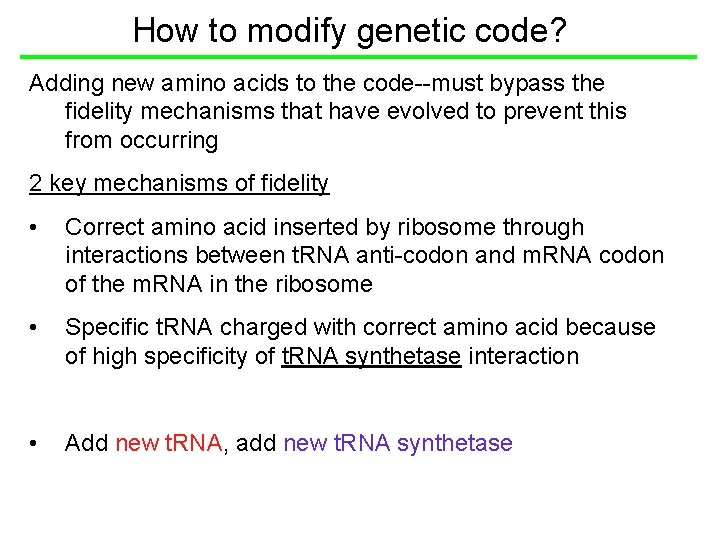
How to modify genetic code? Adding new amino acids to the code--must bypass the fidelity mechanisms that have evolved to prevent this from occurring 2 key mechanisms of fidelity • Correct amino acid inserted by ribosome through interactions between t. RNA anti-codon and m. RNA codon of the m. RNA in the ribosome • Specific t. RNA charged with correct amino acid because of high specificity of t. RNA synthetase interaction • Add new t. RNA, add new t. RNA synthetase

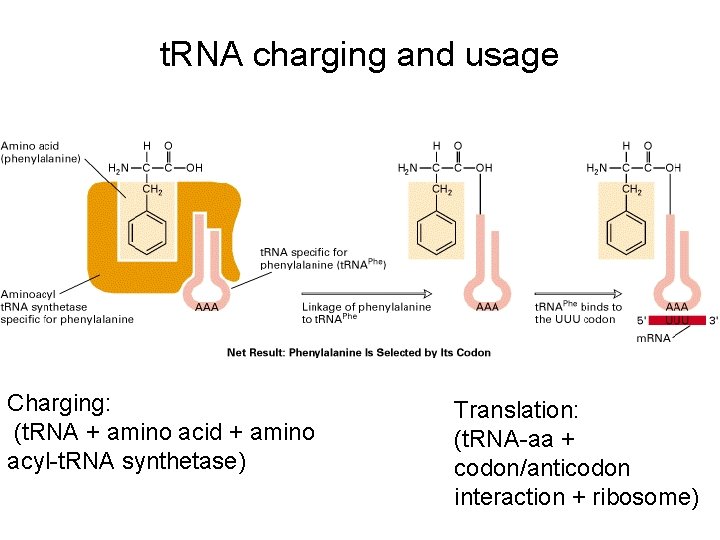
t. RNA charging and usage Charging: (t. RNA + amino acid + amino acyl-t. RNA synthetase) Translation: (t. RNA-aa + codon/anticodon interaction + ribosome)

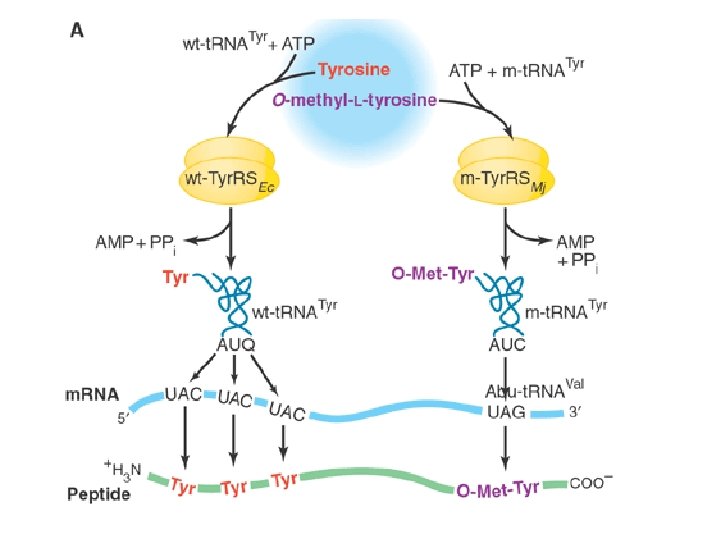
• Chose t. RNAtyr, and the t. RNAtyr synthetase (m. Tyr. RS) from an archaean (M. jannaschii)--no cross-reactivity with E. coli t. RNAtyr and synthetase • Mutate m-t. RNAtyr to recognize stop codon (UAG) on m. RNA • Mutate m-Tyr. RS at 5 positions near the tyrosine binding site by doped oligonucleotide random mutagenesis • Obtain mutants that can insert O-methyl-L-tyrosine at any UAG codon


Outcome • Strategy allows site specific insertion of new amino acid--just design protein to have UAG stop codon where you’d like the new amino acid to go • Transform engineered E. coli with plasmid containing the engineered gene • Feed cells O-methyl tyrosine to get synthesis of full length gene

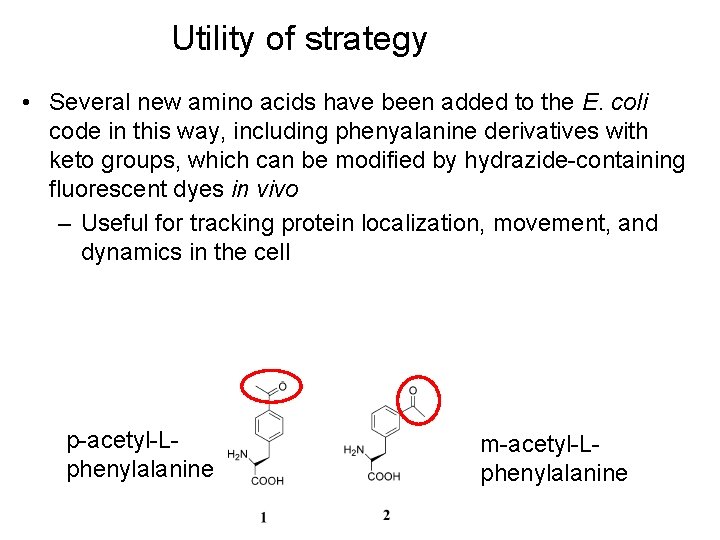
Utility of strategy • Several new amino acids have been added to the E. coli code in this way, including phenyalanine derivatives with keto groups, which can be modified by hydrazide-containing fluorescent dyes in vivo – Useful for tracking protein localization, movement, and dynamics in the cell p-acetyl-Lphenylalanine m-acetyl-Lphenylalanine

Some questions: • What are the consequences for the cell with an expanded code? • Do new amino acids confer any kind of evolutionary advantage to organisms that have them? (assuming they get a ready supply of the new amino acid…) • Why do cells have/need 3 stop codons? ?