Structure of an HIV gp 120 envelope glycoprotein








- Slides: 35

Structure of an HIV gp 120 envelope glycoprotein in complex with the CD 4 receptor and a neutralizing human antibody. Kevin Paiz-Ramirez Janelle N. Ruiz Ryan Willhite Angela Garibaldi Biology 398. 01 Professor Kam D. Dahlquist, Ph. D. Department of Biology Loyola Marymount University March 2 nd, 2009

Outline • Structure Determination • Purpose of Study • Methods • • • Electron Density in Phe 43 Cavity Interfacial cavities Antibody Interface Chemokine Receptor site Oligomer & gp 41 interactoin Conformational changes Viral evasion & Immune response Mechanistic implication for virus entry References

Exploring HIV-1 Structure • Entry of HIV involves a sequential interaction of – the envelope glycoprotein (gp 120) – CD 4 glycoprotein – chemokine receptor (primary receptor) • CD 4 i (antibodies that block gp 120 -CD 4 complexes to the chemokine receptor) • CCR 5 and CXCR 4 for HIV-1 (secondary receptors) • CD 4 binding induces conformational changes in the gp 120 • Entry of HIV is mediated by envelope glycoproteins • 5 variable regions • Variable and non variable regions are glycosylated • V 3 loop determines specificity

Exploring HIV-1 Structure • Gp 41 (transmembrane coat proteins) variants found in all enveloped viruses – N-terminal fusion peptides which participate in membrane fusion • Enveloped viruses tend to be characteristic in entry • Direct membrane penetration (HIV) • HIV causes destruction of CD 4 T cells which ultimately leads to AIDS.

Purpose of Study • Gp 120 glycoprotein has important role in – receptor binding – interactions with neutralizing antibodies • Information about the gp 120 structure is important for understanding HIV infection • Assist in designing therapeutic strategies. • Overall purpose is to observe the mechanism of HIV entry and intervene

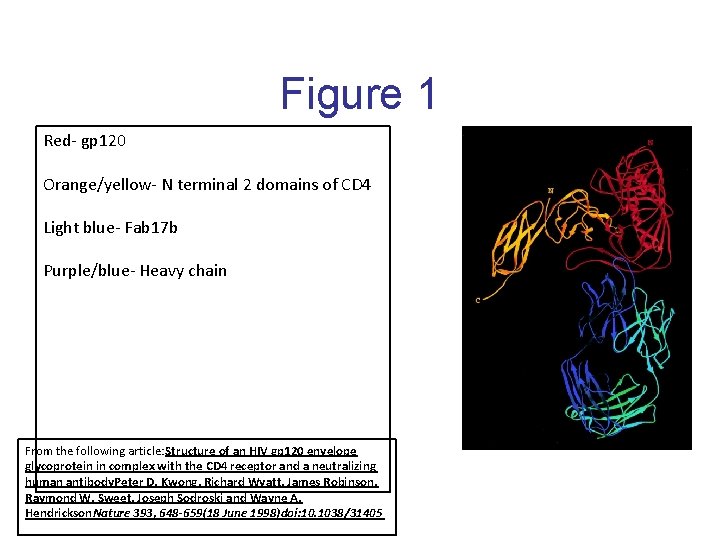
Figure 1 Red- gp 120 Orange/yellow- N terminal 2 domains of CD 4 Light blue- Fab 17 b Purple/blue- Heavy chain From the following article: Structure of an HIV gp 120 envelope glycoprotein in complex with the CD 4 receptor and a neutralizing human antibody. Peter D. Kwong, Richard Wyatt, James Robinson, Raymond W. Sweet, Joseph Sodroski and Wayne A. Hendrickson. Nature 393, 648 -659(18 June 1998)doi: 10. 1038/31405

Determining The Structure • Devised a crystallization strategy that modified the protein surface • Obtained crystals of – a ternary complex composed of a truncated form of gp 120 – the N-terminal two domains (DID 2) of CD 4 – Fab from the human neutralizing monoclonal antibody 17 • The ternary structure was solved by – combinations of molecular replacement – isomorphous replacement – density modification techniques

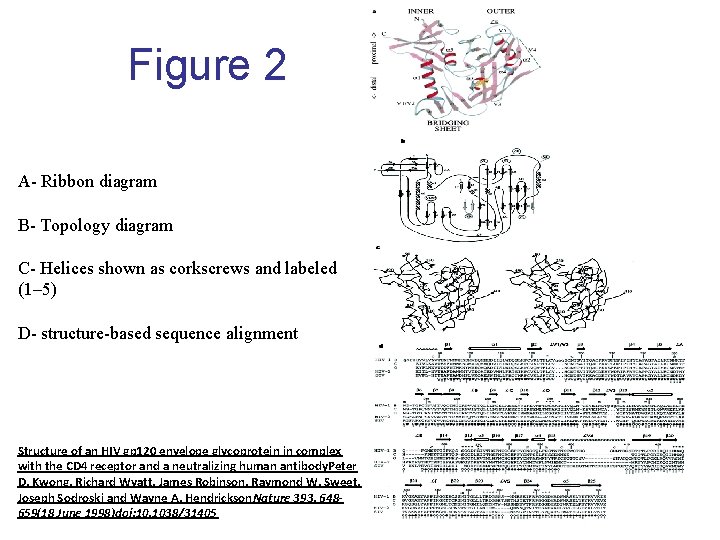
Figure 2 A- Ribbon diagram B- Topology diagram C- Helices shown as corkscrews and labeled (1– 5) D- structure-based sequence alignment Structure of an HIV gp 120 envelope glycoprotein in complex with the CD 4 receptor and a neutralizing human antibody. Peter D. Kwong, Richard Wyatt, James Robinson, Raymond W. Sweet, Joseph Sodroski and Wayne A. Hendrickson. Nature 393, 648659(18 June 1998)doi: 10. 1038/31405

Methods of Determination • Crystallization by modifying protein surface • Deglycosylation of gp 120 variants • Molecular replacement • Isomorphous replacement • Density modification

Crystallization • Crystallize protein to cross-section of 30 -40 um • Measure diffraction patterns with Bragg’s Law to determine space between electrons/atoms (2 d model) – Originally crystals diffracted more than 2 A, but were able to reduce the limit to 2. 5 A

Multiple Isomorphous Replacement • Electrons oscillate during diffraction, must phase them to find electron distribution (location) by inclusion of evenly spaced heavy metals/atom compounds • Tried over 20 different heavy-atom solutions • Isomorphism highest between K 3 Ir. Cl 6, and K 2 Os. Cl 6 and Native form.

Molecular Replacement • Used Fourier Analysis (mathematical method) to determine 3 d pattern of these heavy-atoms (Electron Density Model) • K 3 Ir. Cl 6 modelled as 9 partially occupied sites (2 sites of occupancy) – Poor data quality, small isomorphous differences – K 2 Os. Cl 6 (4 sites of occupancy), highest site at same as 2 nd highest for K 3 Ir. Cl 6

Density Modification to Improve Electron Density Model 1. Correlations in region internal to domain 1 of CD 4 between experimental electron density and calculated model 2. Linkage of unmodeled density (PRISM program) 3. Recipricol-space averaging of the PRISM modeled density 4. Real-space model subtraction (XPLOR) 5. Solvent flattening 6. Histrogram matching 7. Negative-Density Truncation

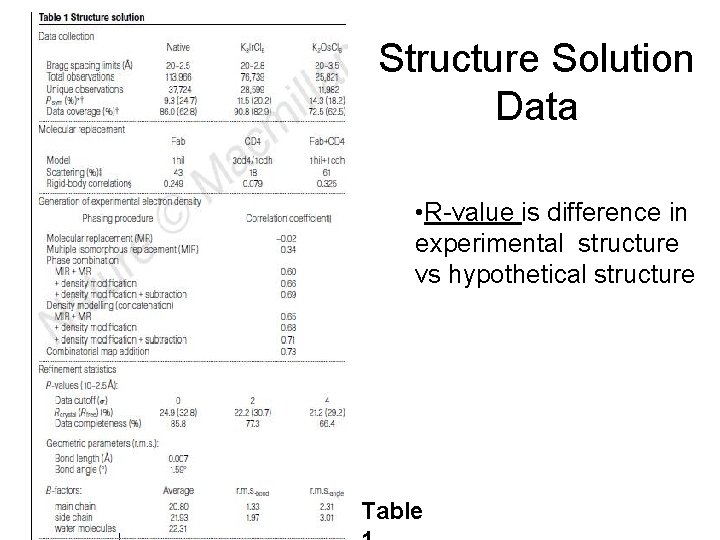
Structure Solution Data • R-value is difference in experimental structure vs hypothetical structure Table

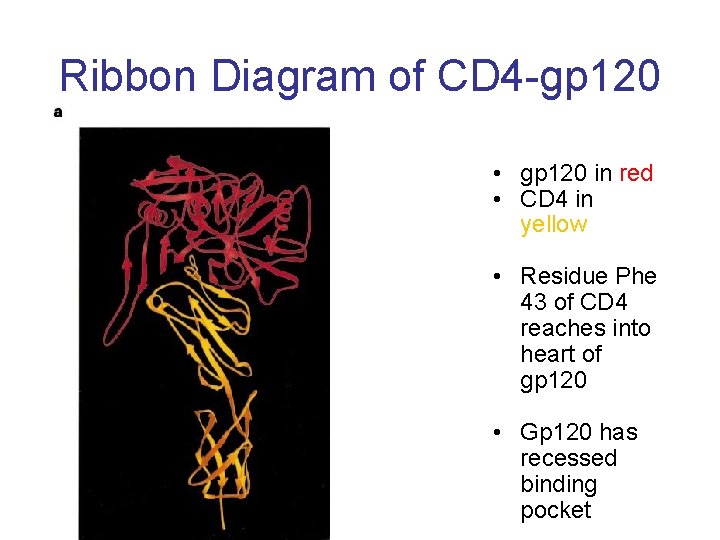
Ribbon Diagram of CD 4 -gp 120 • gp 120 in red • CD 4 in yellow • Residue Phe 43 of CD 4 reaches into heart of gp 120 • Gp 120 has recessed binding pocket

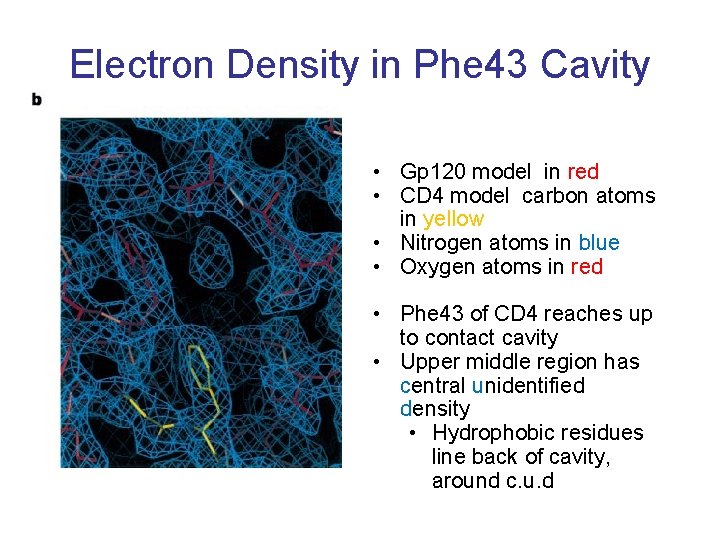
Electron Density in Phe 43 Cavity • Gp 120 model in red • CD 4 model carbon atoms in yellow • Nitrogen atoms in blue • Oxygen atoms in red • Phe 43 of CD 4 reaches up to contact cavity • Upper middle region has central unidentified density • Hydrophobic residues line back of cavity, around c. u. d

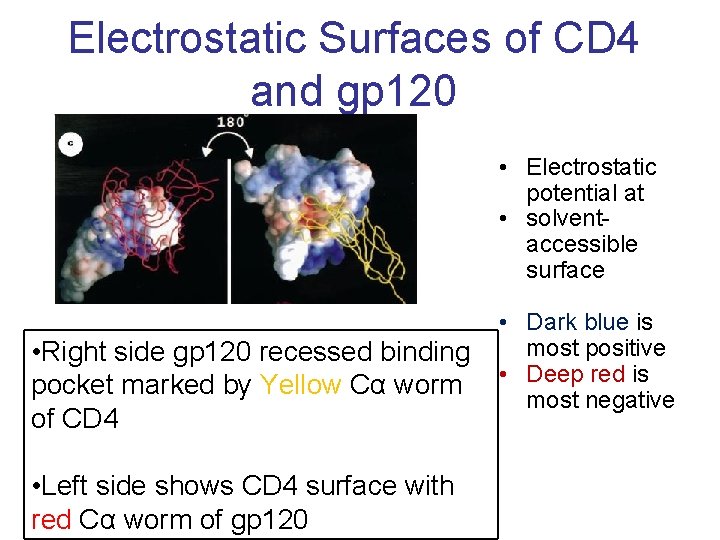
Electrostatic Surfaces of CD 4 and gp 120 • Electrostatic potential at • solventaccessible surface • Right side gp 120 recessed binding pocket marked by Yellow Cα worm of CD 4 • Left side shows CD 4 surface with red Cα worm of gp 120 • Dark blue is most positive • Deep red is most negative

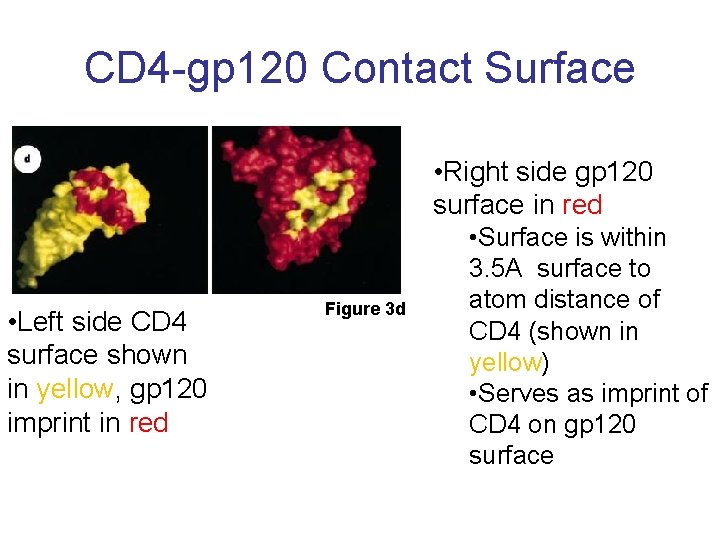
CD 4 -gp 120 Contact Surface • Right side gp 120 surface in red • Left side CD 4 surface shown in yellow, gp 120 imprint in red Figure 3 d • Surface is within 3. 5 A surface to atom distance of CD 4 (shown in yellow) • Serves as imprint of CD 4 on gp 120 surface

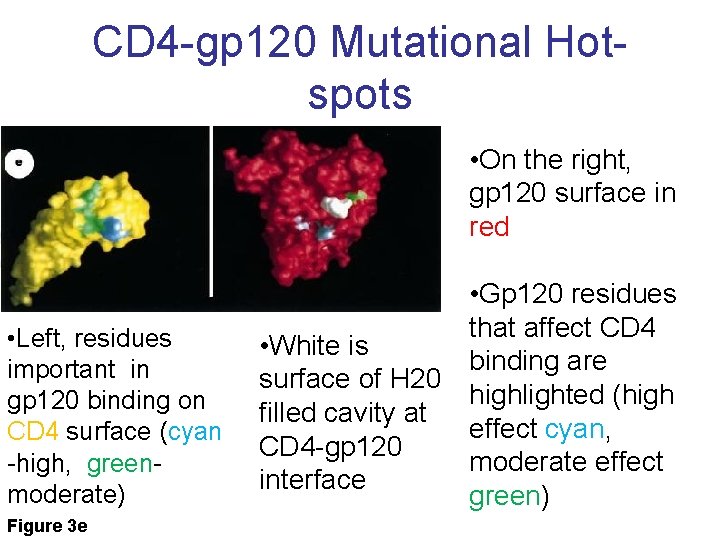
CD 4 -gp 120 Mutational Hotspots • On the right, gp 120 surface in red • Left, residues important in gp 120 binding on CD 4 surface (cyan -high, greenmoderate) Figure 3 e • Gp 120 residues that affect CD 4 • White is binding are surface of H 20 highlighted (high filled cavity at effect cyan, CD 4 -gp 120 moderate effect interface green)

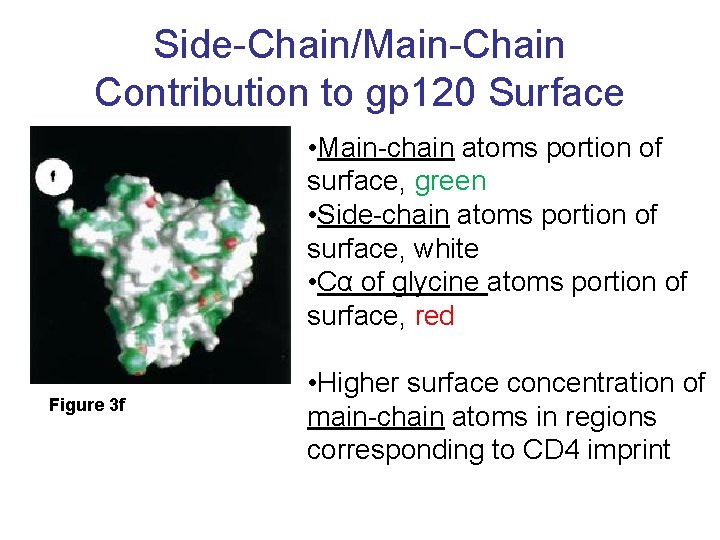
Side-Chain/Main-Chain Contribution to gp 120 Surface • Main-chain atoms portion of surface, green • Side-chain atoms portion of surface, white • Cα of glycine atoms portion of surface, red Figure 3 f • Higher surface concentration of main-chain atoms in regions corresponding to CD 4 imprint

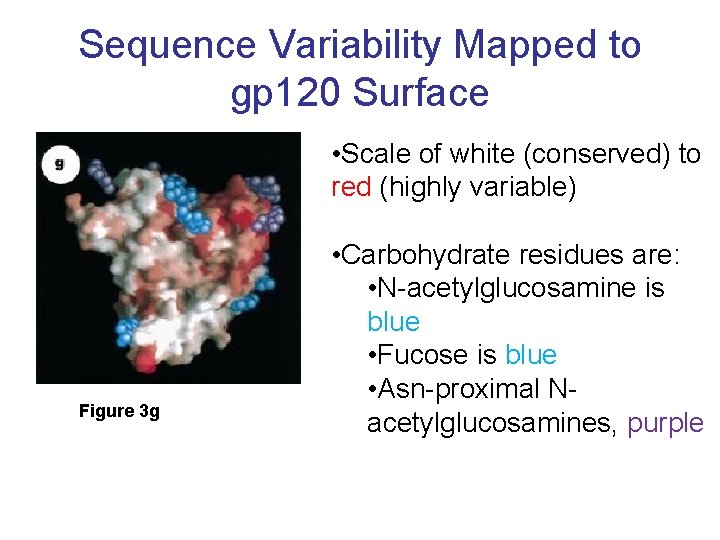
Sequence Variability Mapped to gp 120 Surface • Scale of white (conserved) to red (highly variable) Figure 3 g • Carbohydrate residues are: • N-acetylglucosamine is blue • Fucose is blue • Asn-proximal Nacetylglucosamines, purple

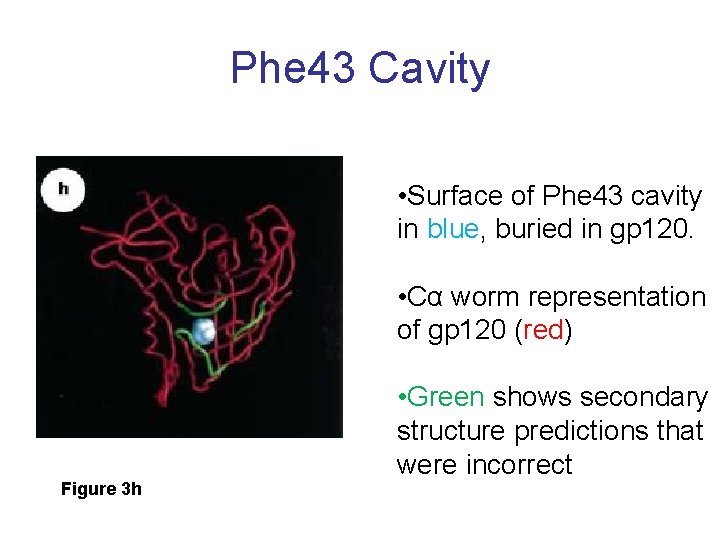
Phe 43 Cavity • Surface of Phe 43 cavity in blue, buried in gp 120. • Cα worm representation of gp 120 (red) Figure 3 h • Green shows secondary structure predictions that were incorrect

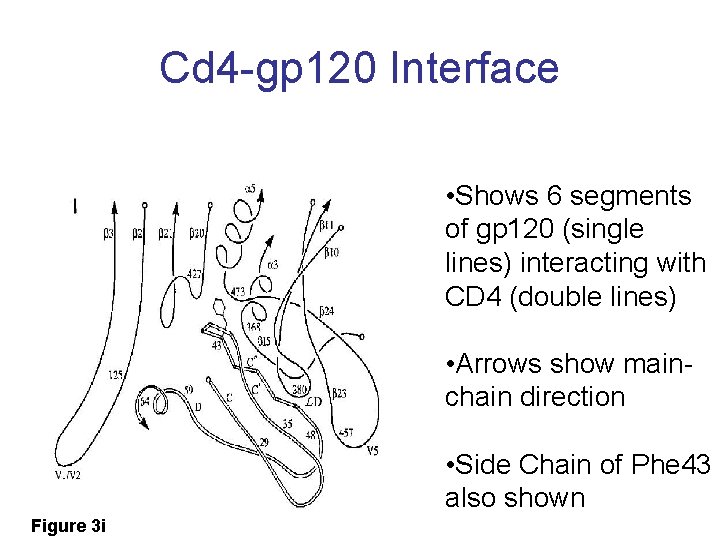
Cd 4 -gp 120 Interface • Shows 6 segments of gp 120 (single lines) interacting with CD 4 (double lines) • Arrows show mainchain direction • Side Chain of Phe 43 also shown Figure 3 i

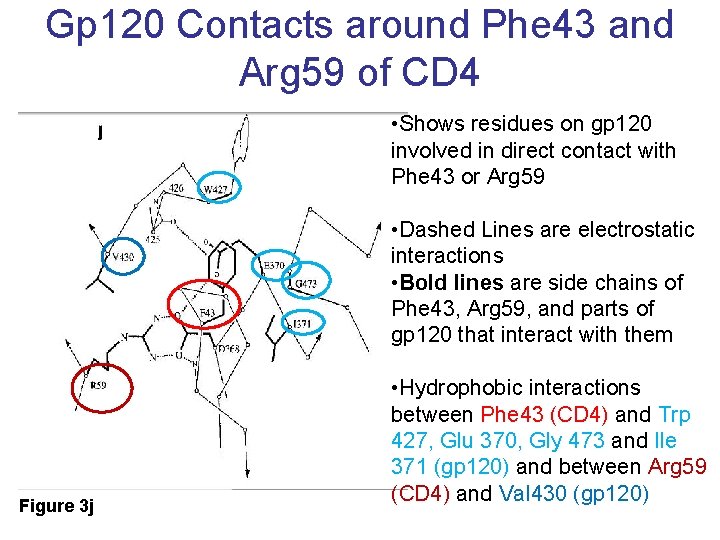
Gp 120 Contacts around Phe 43 and Arg 59 of CD 4 • Shows residues on gp 120 involved in direct contact with Phe 43 or Arg 59 • Dashed Lines are electrostatic interactions • Bold lines are side chains of Phe 43, Arg 59, and parts of gp 120 that interact with them Figure 3 j • Hydrophobic interactions between Phe 43 (CD 4) and Trp 427, Glu 370, Gly 473 and Ile 371 (gp 120) and between Arg 59 (CD 4) and Val 430 (gp 120)

Interfacial Cavities • Analysis of the surface of the ternary complex revealed topological surface cavities. • The gp 120 -CD 4 interface were unusually large. The cavity served as a water buffer between gp 120 and CD 4. • The residue that line this cavity provide little direct contact to CD 4. This can reduce binding. This can affect the binding of antibodies against the CD 4 binding site. • Despite the unusual cavity laden interface between gp 120 and CD 4 the missing gp 120 loops and termini could not have a role in filling these cavities.

Antibody Interface • Concerning the 17 b antibody, a broadly neutralizing human monoclonal isolated from HIV infected individuals that bind to CD 4 induced gp 120 epitrope, that overlaps into the chemokine receptor binding site. • The 17 b contact surface is very acidic. • Tested against four stranded bridging sheet. Contact suggested that the sheets were necessary for 17 b binding. • The interaction between 17 b and gp 120 involves a hydrophobic central region flanked with periphery by charged regions • There is no direct CD 4 -17 b contact.

Chemokine-Receptor site • The chemokine receptor, CCR 5 overlaps with 17 b, both are induced upon CD 4 binding and both involve highly conserved residues. • The hydrophobic and acidic surface of 17 b heavy chain may mimic the tyrosine rich acidic N-Terminal region of CCR 5.

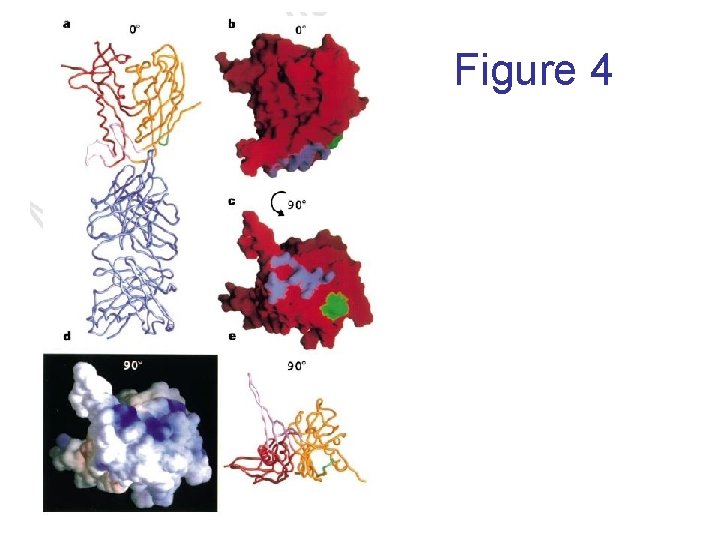
Figure 4

Oligomer and gp 41 interaction • gp 120 exist as a trimeric complex with gp 41 on the virion surface. • The N and C termini of full length gp 120 are the most important regions for interaction with the gp 41 glycoprotein. • It was expected that gp 41 interactive regions will extent away from core gp 120 toward the viral membrane that is conserved.

Conformational change in core gp 120 • Evidence for CD 4 -induced conformational change: 1. Structural dilemma with Phe 43 cavity 2. Characteristics of 17 b binding to core gp 120 3. Comparison with theory • Evolutionary algorithms of known sequence variants of gp 120 gives secondary structure predictions with high reliability 4. Phe 43 cavity is the nexus of the CD 4 interface -- lying b/w the inner and outer domain and bridging sheet-without Phe 3 structure might collapse!

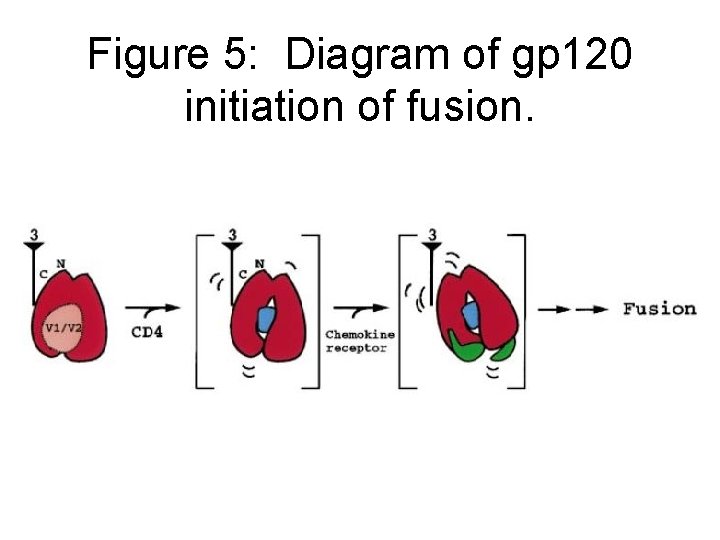
Figure 5: Diagram of gp 120 initiation of fusion.

Viral Evasion of Immune Response • Analysis of antigenic structure of gp 120 shows that most of the envelope protein surface is hidden from humoral immune response by glycosylation and oligomeric blockage • Most neutralizing antibodies access only two surfaces: CD 4 bidning site and chemokine-receptor binding site • Conformational changes in core gp 120 • CD 4 -binding site recognition prevented by: recessed nature of the binding pocket and topographical surface mismatch • Chemokine receptor region recognition prevented by: conformational change; steric blockage; surface mismatch

Mechanistic implications for virus entry • Gp 120 crucial for fusion of HIV to cell surface • Given data presented here, suggest this may be a simple process comprising viral oligomer and two host receptors. • Gp 120 functions in: POSITIONING • Gp 120 function in: TIMING • Entry process initiated by binding of HIV-1 to cellular receptor CD 4 -- orients viral spike • CD 4 binding induces conformational change in gp 120

Mechanistic implications for virus entry • CD 4 binding results in movement of V 2 loop, which partially blocks V 3 loop and CD 4 interacting epitopes • Also creates/stabilizes bridging sheet • CD 4 binding changes V 3 region -- altering exposure of V 3 epitopes • Next step: Interaction of gp 120 -CD 4 complex with chemokine receptor • Binding will move gp 120 closer to target membrane MAIN POINT: Structure of gp 120/CD 4/17 b antibody ternary complex described here revels many , molecular aspects of HIV-1 entry

References • Kwong Peter, Wyatt Richard, Robinson James, Sweets Raymond, Sodroski Joseph, and Wayne Hendrickson. Structure of an HIV gp 120 envelope glycoprotein in complex with the CD 4 receptor and a neutralizing human antibody. Proc Natl Acad Sci USA 1998.