Structure of a typical operon The enzymes that








- Slides: 16





Structure of a typical operon




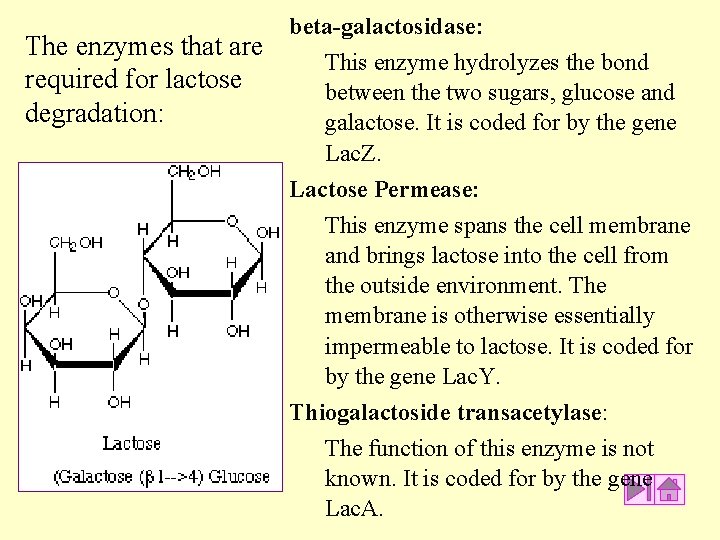
The enzymes that are required for lactose degradation: beta-galactosidase: This enzyme hydrolyzes the bond between the two sugars, glucose and galactose. It is coded for by the gene Lac. Z. Lactose Permease: This enzyme spans the cell membrane and brings lactose into the cell from the outside environment. The membrane is otherwise essentially impermeable to lactose. It is coded for by the gene Lac. Y. Thiogalactoside transacetylase: The function of this enzyme is not known. It is coded for by the gene Lac. A.

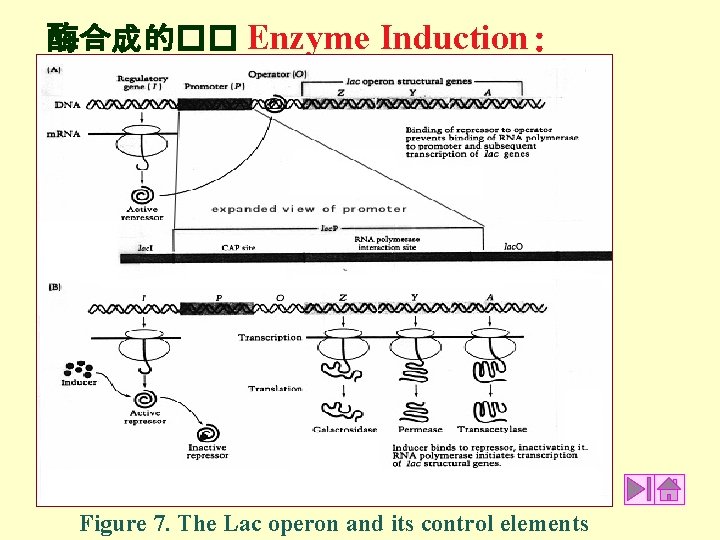
酶合成的�� Enzyme Induction: Figure 7. The Lac operon and its control elements

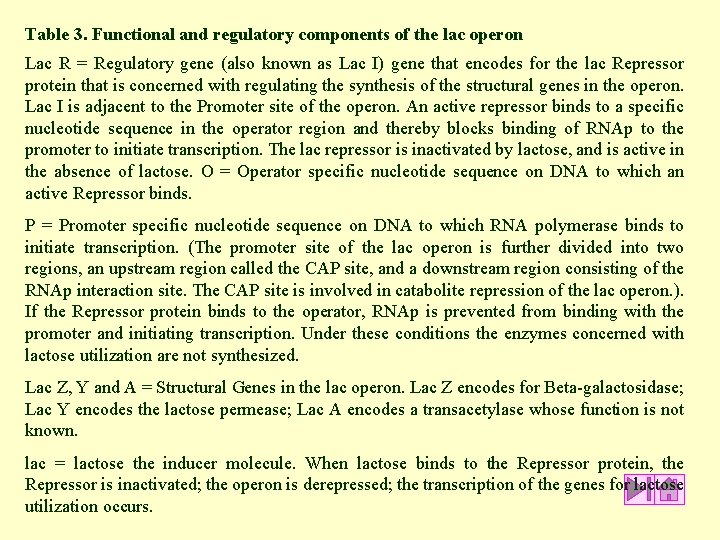
Table 3. Functional and regulatory components of the lac operon Lac R = Regulatory gene (also known as Lac I) gene that encodes for the lac Repressor protein that is concerned with regulating the synthesis of the structural genes in the operon. Lac I is adjacent to the Promoter site of the operon. An active repressor binds to a specific nucleotide sequence in the operator region and thereby blocks binding of RNAp to the promoter to initiate transcription. The lac repressor is inactivated by lactose, and is active in the absence of lactose. O = Operator specific nucleotide sequence on DNA to which an active Repressor binds. P = Promoter specific nucleotide sequence on DNA to which RNA polymerase binds to initiate transcription. (The promoter site of the lac operon is further divided into two regions, an upstream region called the CAP site, and a downstream region consisting of the RNAp interaction site. The CAP site is involved in catabolite repression of the lac operon. ). If the Repressor protein binds to the operator, RNAp is prevented from binding with the promoter and initiating transcription. Under these conditions the enzymes concerned with lactose utilization are not synthesized. Lac Z, Y and A = Structural Genes in the lac operon. Lac Z encodes for Beta-galactosidase; Lac Y encodes the lactose permease; Lac A encodes a transacetylase whose function is not known. lac = lactose the inducer molecule. When lactose binds to the Repressor protein, the Repressor is inactivated; the operon is derepressed; the transcription of the genes for lactose utilization occurs.

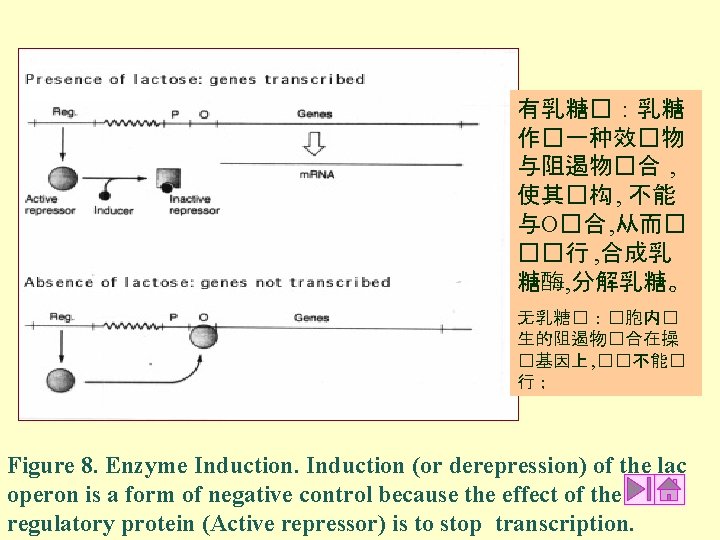
有乳糖�:乳糖 作�一种效�物 与阻遏物�合 , 使其�构 , 不能 与O�合 , 从而� ��行 , 合成乳 糖酶, 分解乳糖。 无乳糖�:�胞内� 生的阻遏物�合在操 �基因上 , ��不能� 行; Figure 8. Enzyme Induction (or derepression) of the lac operon is a form of negative control because the effect of the regulatory protein (Active repressor) is to stop transcription.


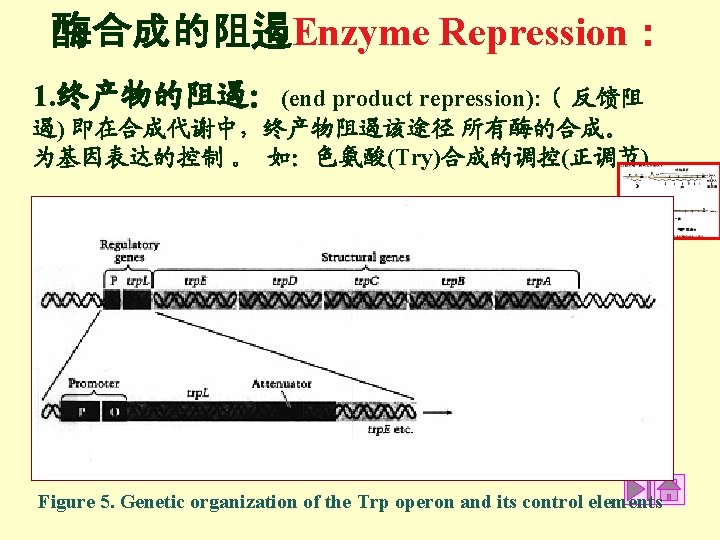
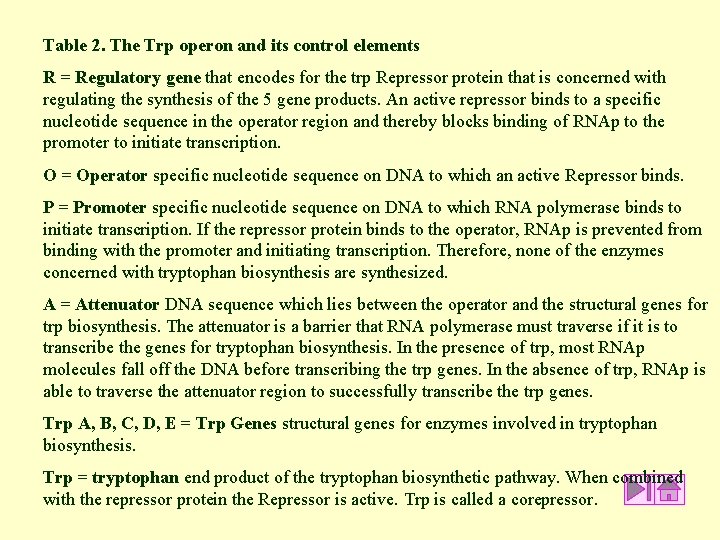
Table 2. The Trp operon and its control elements R = Regulatory gene that encodes for the trp Repressor protein that is concerned with regulating the synthesis of the 5 gene products. An active repressor binds to a specific nucleotide sequence in the operator region and thereby blocks binding of RNAp to the promoter to initiate transcription. O = Operator specific nucleotide sequence on DNA to which an active Repressor binds. P = Promoter specific nucleotide sequence on DNA to which RNA polymerase binds to initiate transcription. If the repressor protein binds to the operator, RNAp is prevented from binding with the promoter and initiating transcription. Therefore, none of the enzymes concerned with tryptophan biosynthesis are synthesized. A = Attenuator DNA sequence which lies between the operator and the structural genes for trp biosynthesis. The attenuator is a barrier that RNA polymerase must traverse if it is to transcribe the genes for tryptophan biosynthesis. In the presence of trp, most RNAp molecules fall off the DNA before transcribing the trp genes. In the absence of trp, RNAp is able to traverse the attenuator region to successfully transcribe the trp genes. Trp A, B, C, D, E = Trp Genes structural genes for enzymes involved in tryptophan biosynthesis. Trp = tryptophan end product of the tryptophan biosynthetic pathway. When combined with the repressor protein the Repressor is active. Trp is called a corepressor.

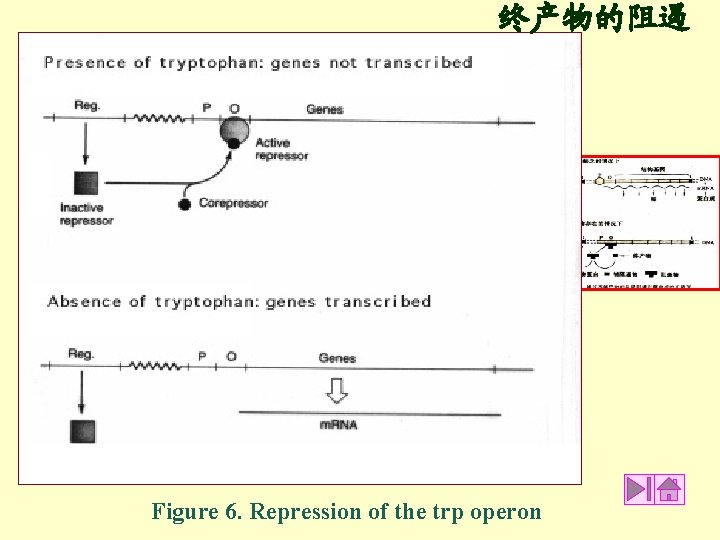
终产物的阻遏 Figure 6. Repression of the trp operon

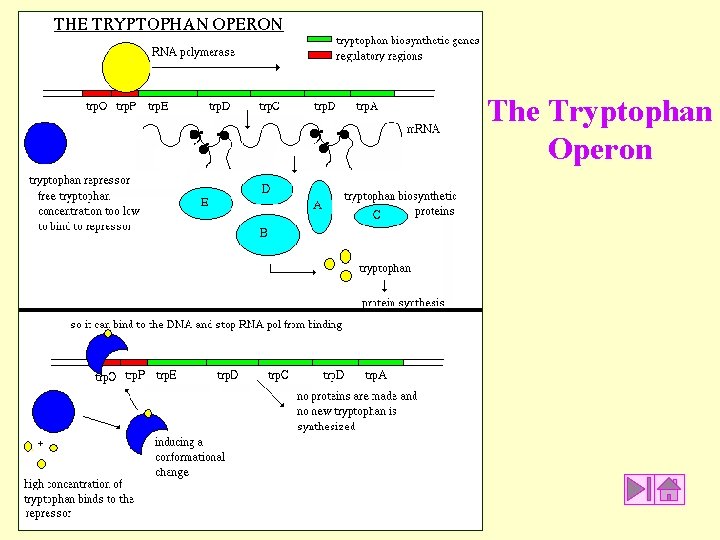
The Tryptophan Operon