Structure determination by NMR principles Data acquisition Spectra
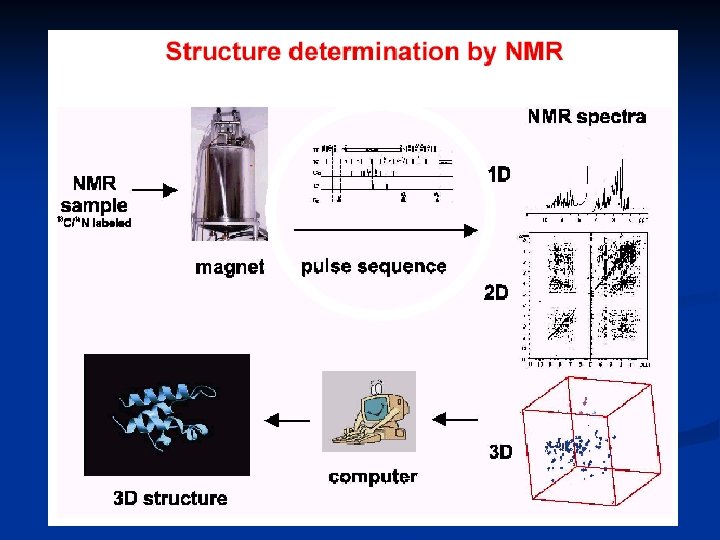
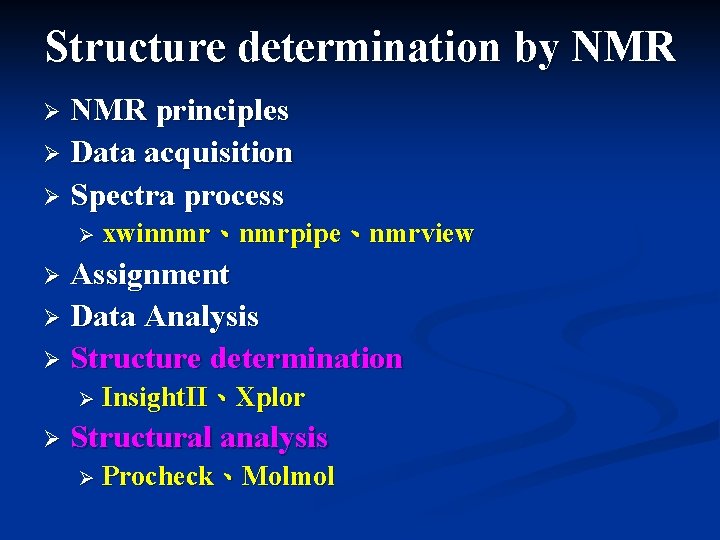
Structure determination by NMR principles Ø Data acquisition Ø Spectra process Ø Ø xwinnmr、nmrpipe、nmrview Assignment Ø Data Analysis Ø Structure determination Ø Ø Insight. II、Xplor Ø Structural analysis Ø Procheck、Molmol

Structure determination and analysis Steps : 1. How to Run X-PLOR 2. Starting Structure 3. PSF file

Structure determination and analysis 1. How to Run X-PLOR程式在各個主機的路徑如下: Hybrid : /disk 2/programs/xplor BTK : /disk 3/programs/xplor Biosym : /disk 2/programs/xplor

Structure determination and analysis 欲執行X-PLOR� 只需在自己帳號下的. cshrc file 內加入下列 script。 以hybrid為例: alias Xplor '/disk 2/programs/xplor/Xplor. exe' 在 unix 環境下鍵入 Xplor <input_filename> output_filename &

Structure determination and analysis 在 unix 環境下鍵入 Xplor <input_filename> output_filename & input_filename : 執行檔,這些檔案通稱為 input file,其檔名皆 為 *. inp。(Ex. sa. inp、psf. inp、average. inp、refine. inp…) output_filename : X-PLOR可將 input file 執行的狀況寫入另一 檔案內,以便了解問題所在,此檔案稱為output file� 我習 慣命名為 *. out & : 此記號可將此次作業轉為幕後執行,並給你執行序號。 終止幕後執行的指令是:kill 執行序號

Structure determination and analysis 2. Starting Structure Insight 97

Structure determination and analysis 2. Starting Structure – way 1 n 1. 先按住左上角msi的mark, 選 Biopolymer, 便出現如上 圖之視窗 n 2. Residue --> Append Enter the Molecular Name choose the Residue (from N-terminal to C-terminal) choose the way to append (we choose "Extended". )

Structure determination and analysis n 3. Protein Cap C-terminus COO- 、N-terminus NH 3 <註> 給N和C端正確的帶電性及原子數 n 4. Modify Hydrogens Set_p. H = 7. 0 Capping_Mode Charged <註> 給Sidechain正確的帶電性及原子數 n 5. Molecule Put File Type XPLOR Mulecule Name

Structure determination and analysis 2. Starting Structure – way 2 n 文字指令指令內容:build. log n 執行方式:File Source File ,choose build. log

Structure determination and analysis 2. Starting Structure – way 2 n build. log #Running: /disk 2/msi/970/irix 6 m 3/biosym_exe/insight. II #Log Created: Tue May 26 17: 56: 06 1998 # #Insight II Version 97. 0 - Molecular Modeling System m: Biopolymer m: Append Residue test ALA Extended r: Append Residue test LEU Extended r: Append Residue test SER Extended m: Cap Protein TEST C_Terminus COO- N_Terminus NH 3 m: Hydrogens TEST set_p. H 7 -Lone_Pairs Charged -Assign_IUPAC_Names m: Put Molecule XPLOR TEST test. pdb -Transformed -XPLOR_PSF

Structure determination and analysis 3. PSF file n PDB File :定義蛋白質結構的空間座標 n PSF File (Protein Structure File:記錄所有原子的電性, 質量 和各個鍵結及鍵角等資料。 指令:Xplor <psf. inp> psf. out &
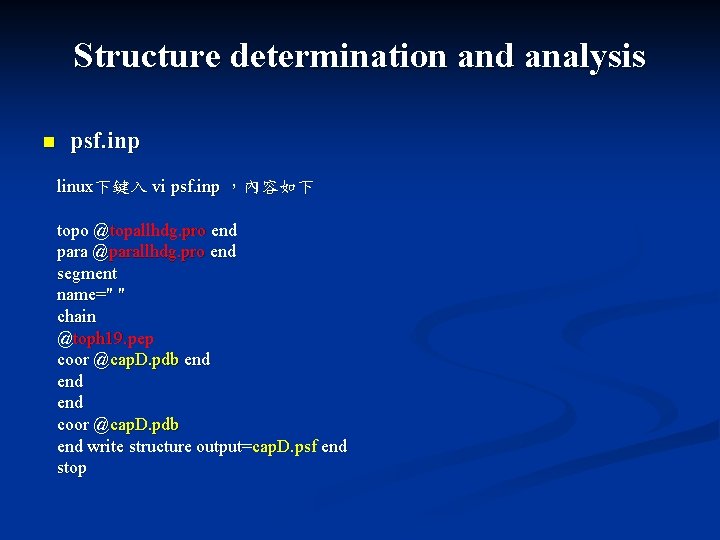
Structure determination and analysis n psf. inp linux下鍵入 vi psf. inp ,內容如下 topo @topallhdg. pro end para @parallhdg. pro end segment name=" " chain @toph 19. pep coor @cap. D. pdb end end coor @cap. D. pdb end write structure output=cap. D. psf end stop
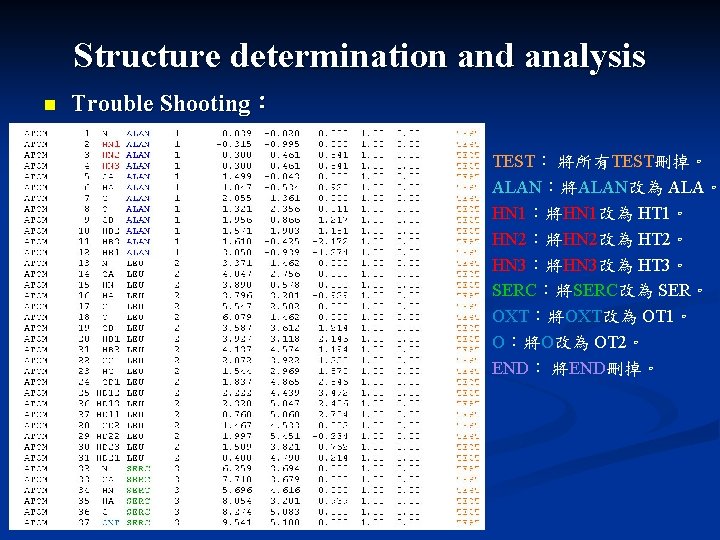
Structure determination and analysis n Trouble Shooting: TEST: 將所有TEST刪掉。 ALAN:將ALAN改為 ALA。 HN 1:將HN 1改為 HT 1。 HN 2:將HN 2改為 HT 2。 HN 3:將HN 3改為 HT 3。 SERC:將SERC改為 SER。 OXT:將OXT改為 OT 1。 O:將O改為 OT 2。 END: 將END刪掉。
- Slides: 14