Structure and Replication of DNA Outline DNA as







- Slides: 69

Structure and Replication of DNA

Outline • DNA as the genetic material (16. 2, 16. 3, 16. 4) • Watson-Crick model of DNA Structure (16. 5, 16. 7, 16. 6, 16. 8, ) • Semiconservative model of DNA replication (16. 9, 16. 10, 16. 11, 16. 12, 16. 13, 16. 14, 16. 15, 16. 16, 16. 17, ) • Repair of Damaged DNA (16. 18, 16. 19) • Telomerase extension of chromosome ends (16. 20) • Chromatin (16. 21)

Are Genes Composed of DNA or Protein? • DNA – Only four nucleotides • thought to have monotonous structure • Protein – 20 different amino acids – greater potential variation – More protein in chromosomes than DNA

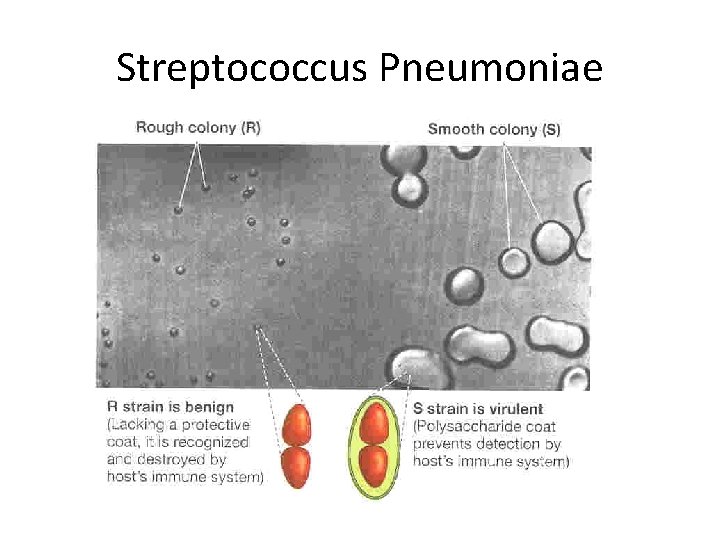
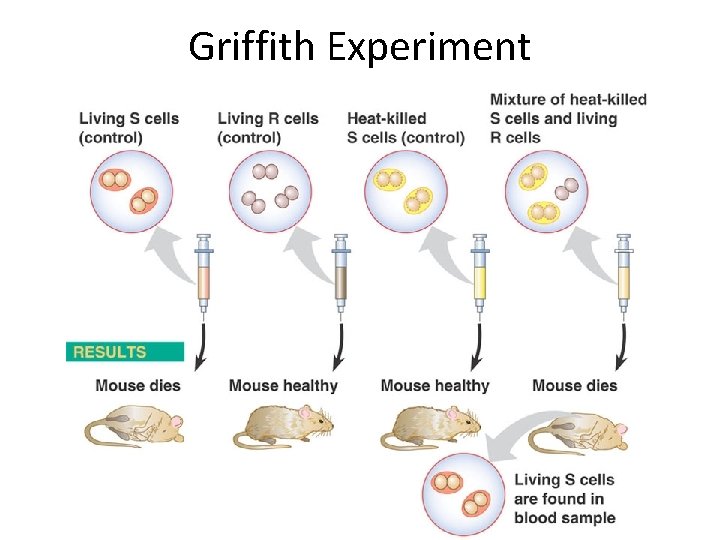
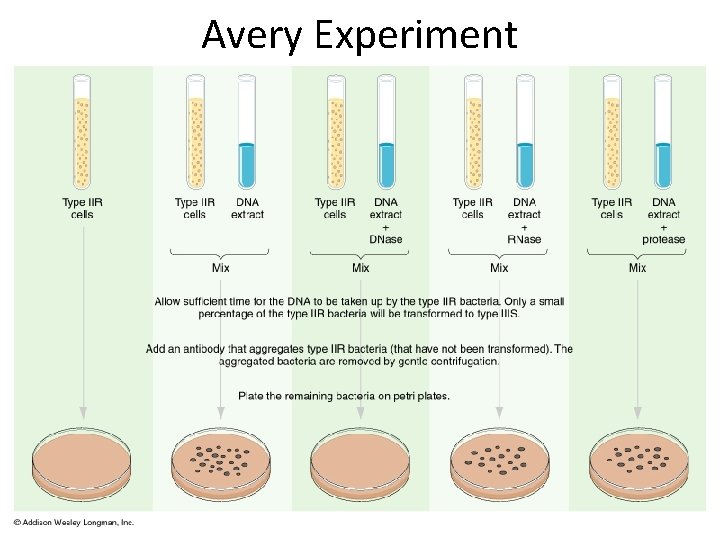
Bacterial Transformation Experiments Fredrick Griffith (1928) –demonstrate the existence of “Transforming Principle, ” a substance able to confer a heritable phenotype from one strain of bacteria to another. Avery Mac. Leod and Mc. Carty – determine the transforming principle was DNA.

Streptococcus Pneumoniae

Griffith Experiment

Avery Experiment

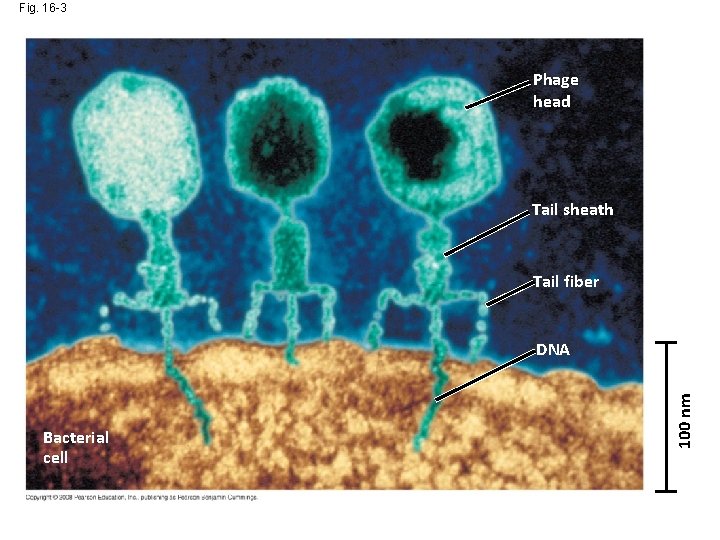
Fig. 16 -3 Phage head Tail sheath Tail fiber Bacterial cell 100 nm DNA

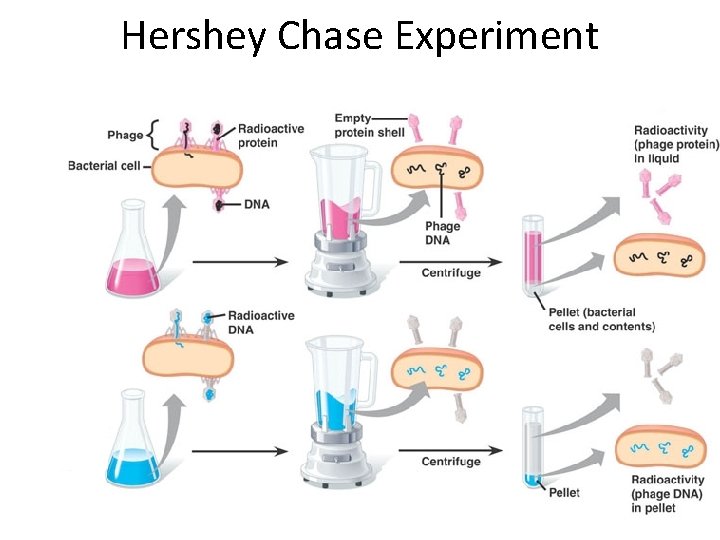
Hershey Chase Experiment

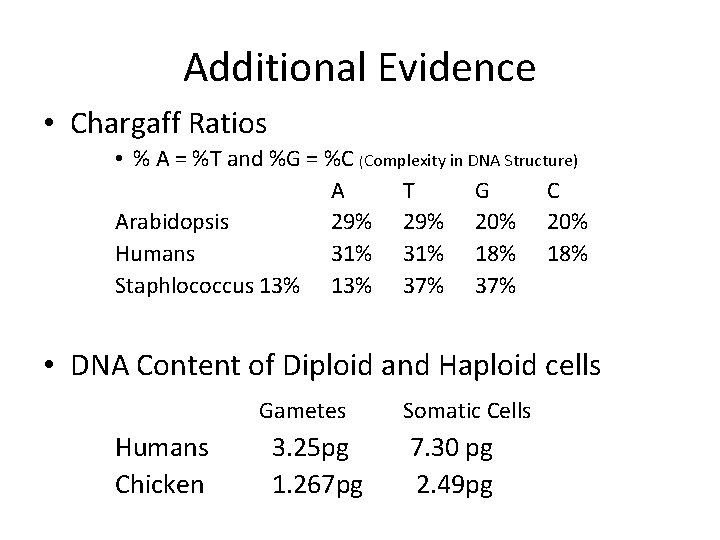
Additional Evidence • Chargaff Ratios • % A = %T and %G = %C (Complexity in DNA Structure) A T G C Arabidopsis 29% 20% Humans 31% 18% Staphlococcus 13% 37% • DNA Content of Diploid and Haploid cells Gametes Humans Chicken 3. 25 pg 1. 267 pg Somatic Cells 7. 30 pg 2. 49 pg

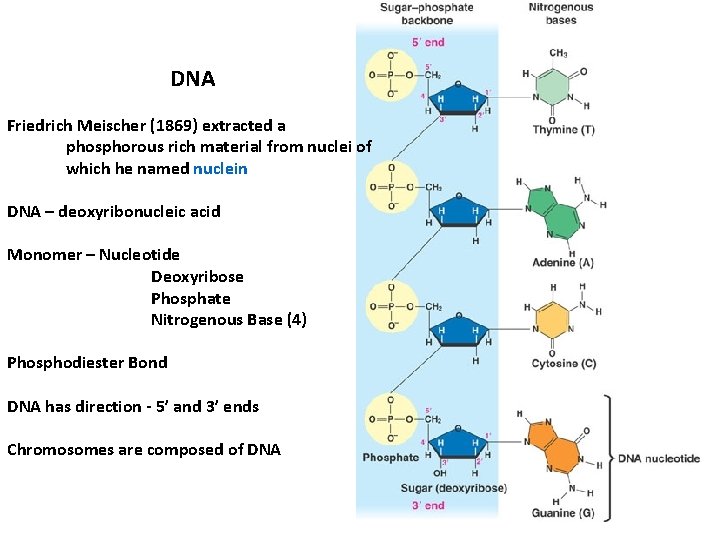
DNA Friedrich Meischer (1869) extracted a phosphorous rich material from nuclei of which he named nuclein DNA – deoxyribonucleic acid Monomer – Nucleotide Deoxyribose Phosphate Nitrogenous Base (4) Phosphodiester Bond DNA has direction - 5’ and 3’ ends Chromosomes are composed of DNA

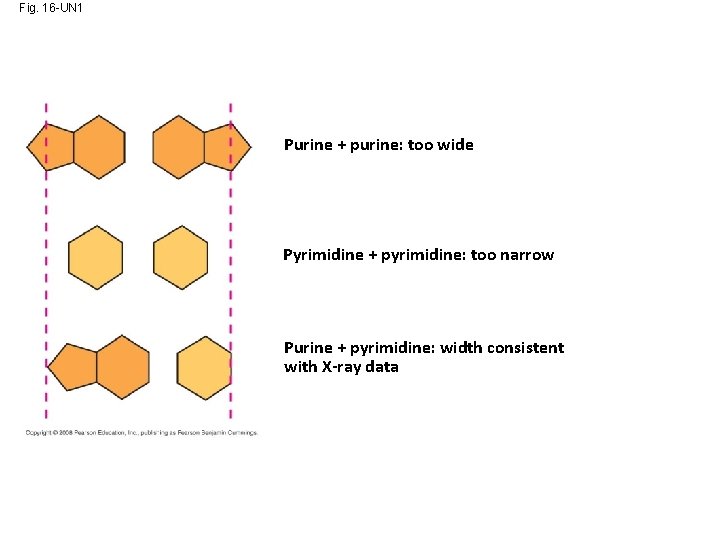
Fig. 16 -UN 1 Purine + purine: too wide Pyrimidine + pyrimidine: too narrow Purine + pyrimidine: width consistent with X-ray data

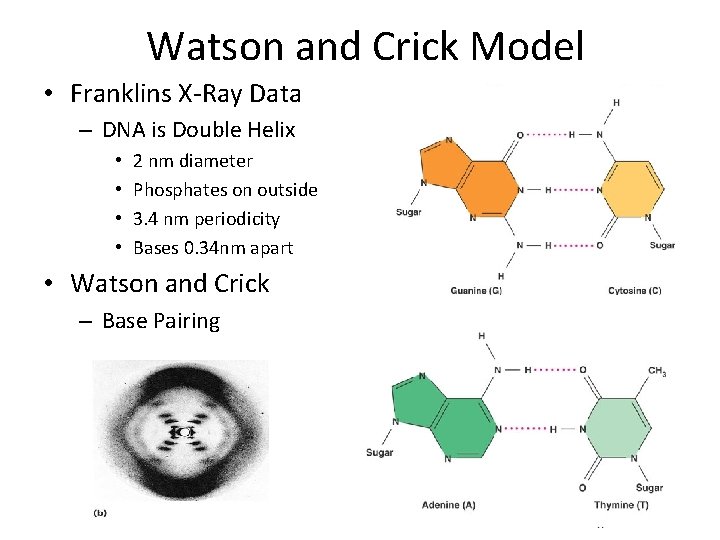
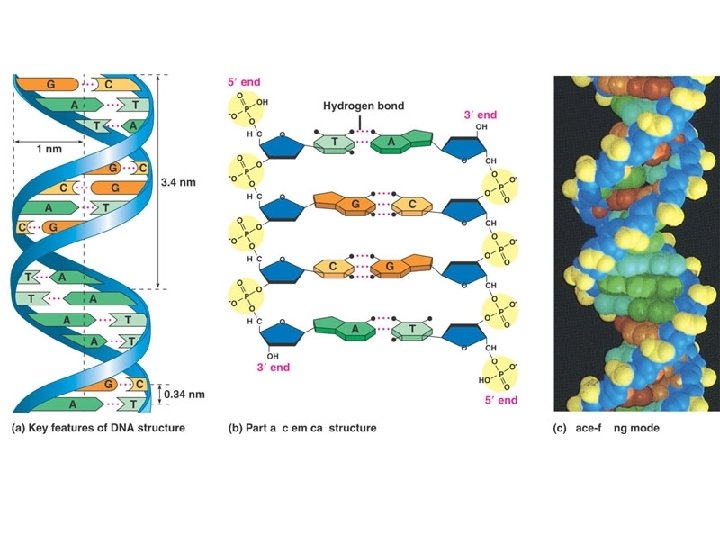
Watson and Crick Model • Franklins X-Ray Data – DNA is Double Helix • • 2 nm diameter Phosphates on outside 3. 4 nm periodicity Bases 0. 34 nm apart • Watson and Crick – Base Pairing


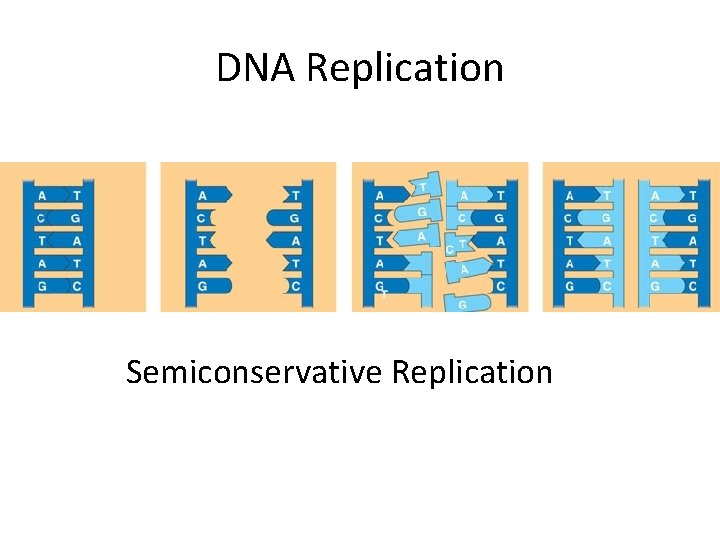
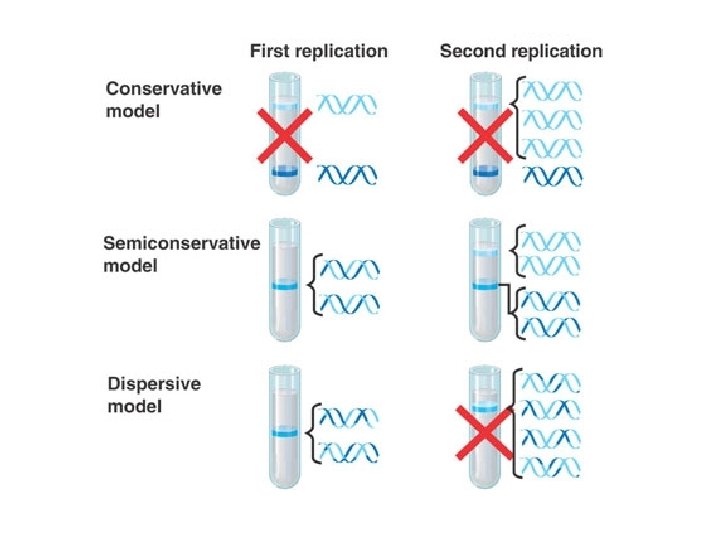
DNA Replication Semiconservative Replication

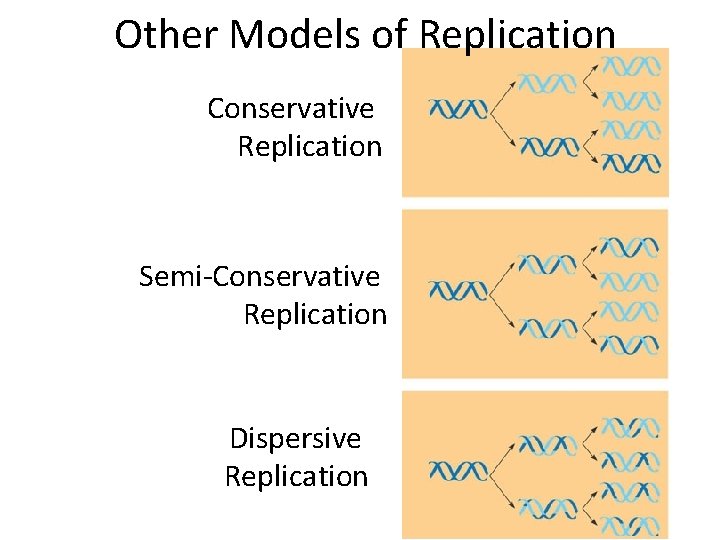
Other Models of Replication Conservative Replication Semi-Conservative Replication Dispersive Replication

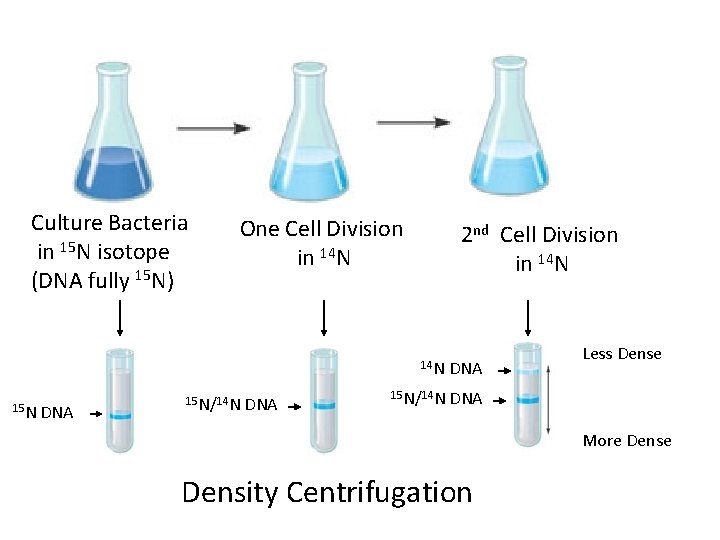
Culture Bacteria in 15 N isotope (DNA fully 15 N) 15 N DNA One Cell Division in 14 N 15 N/14 N DNA 2 nd Cell Division in 14 N DNA 15 N/14 N DNA Less Dense More Density Centrifugation


DNA Replication: A Closer Look • The copying of DNA is remarkable in its speed and accuracy • More than a dozen enzymes and other proteins participate in DNA replication Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

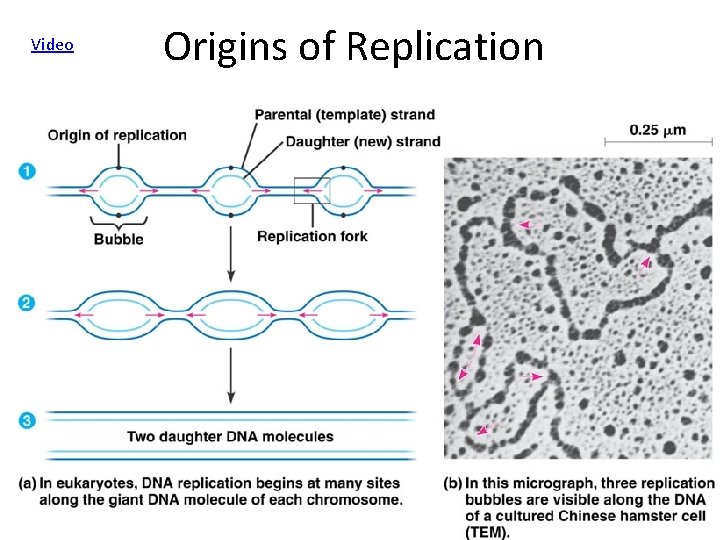
Video Origins of Replication

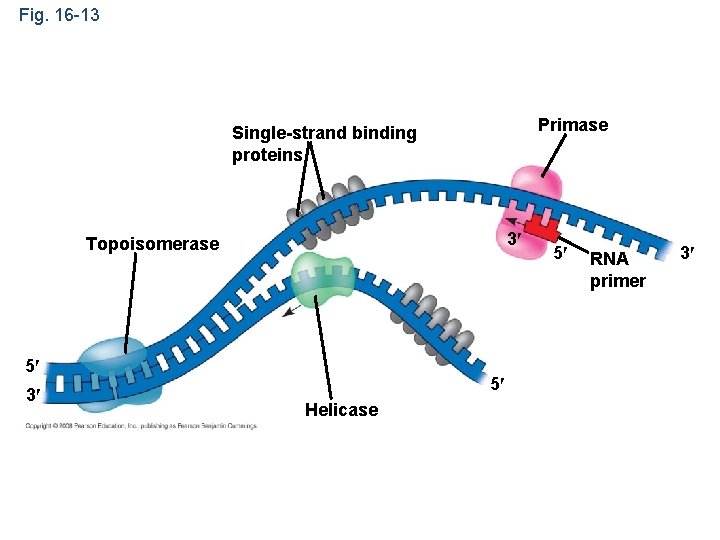
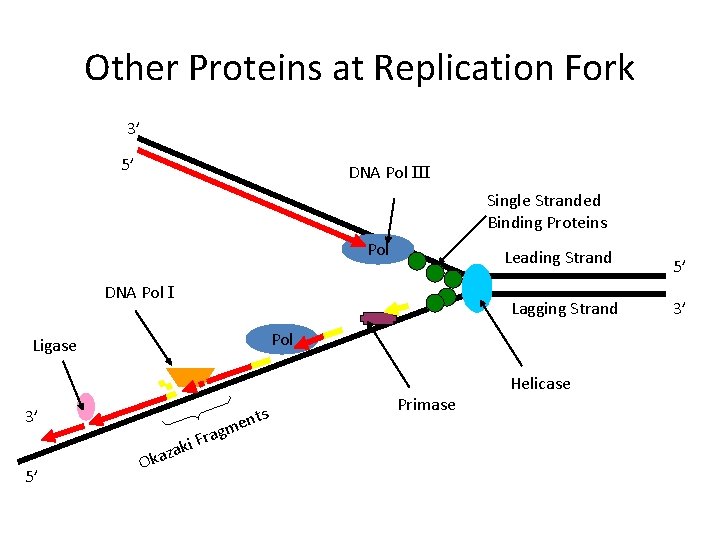
• At the end of each replication bubble is a replication fork, a Y-shaped region where new DNA strands are elongating • Helicases are enzymes that untwist the double helix at the replication forks • Single-strand binding protein binds to and stabilizes single-stranded DNA until it can be used as a template • Topoisomerase corrects “overwinding” ahead of replication forks by breaking, swiveling, and rejoining DNA strands Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Fig. 16 -13 Primase Single-strand binding proteins 3 Topoisomerase 5 3 5 Helicase 5 RNA primer 3


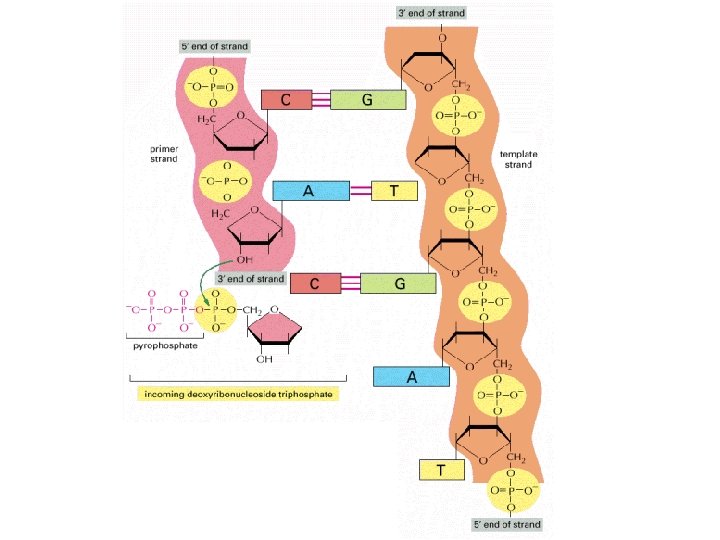
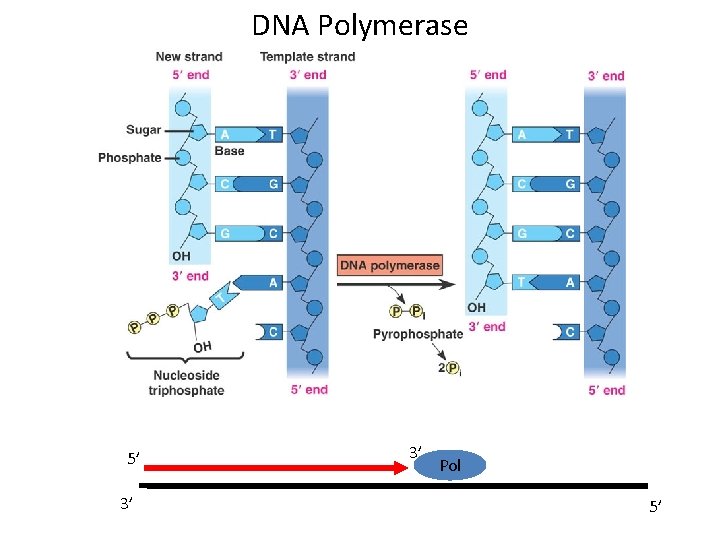
DNA Polymerase 5’ 3’ 3’ Pol 5’

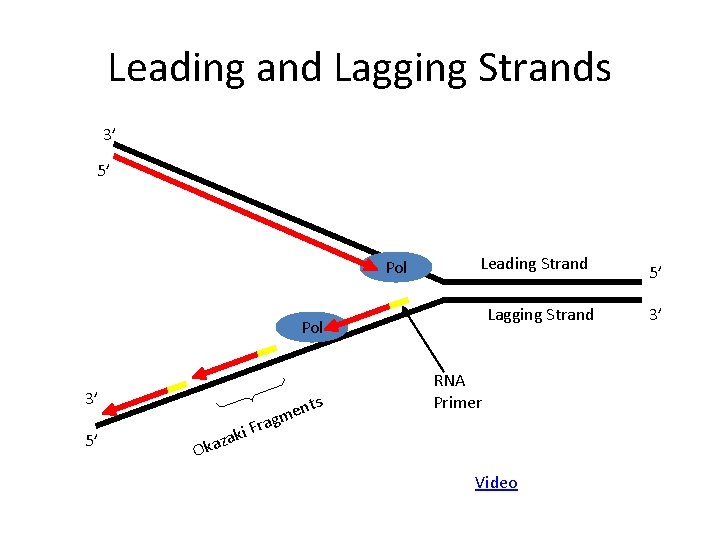
Leading and Lagging Strands 3’ 5’ Pol Leading Strand Lagging Strand Pol 3’ 5’ ts en m g i Fra RNA Primer k za a k O Video 5’ 3’

Other Proteins at Replication Fork 3’ 5’ DNA Pol III Single Stranded Binding Proteins Pol Leading Strand DNA Pol I Lagging Strand Pol Ligase 3’ 5’ za a k O rag F i k m s ent Primase Helicase 5’ 3’

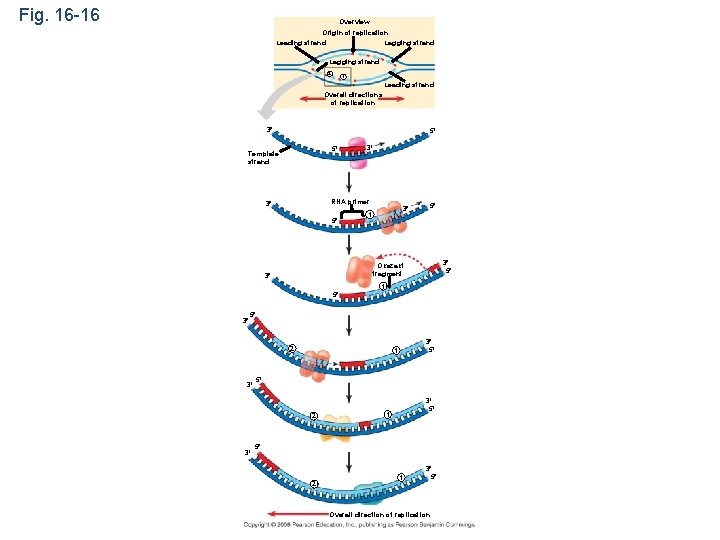
Fig. 16 -16 Overview Origin of replication Lagging strand Leading strand Lagging strand 2 1 Leading strand Overall directions of replication 3 5 5 Template strand 3 RNA primer 3 5 3 1 5 3 5 Okazaki fragment 3 1 5 3 5 2 3 3 5 1 5 2 1 3 5 Overall direction of replication

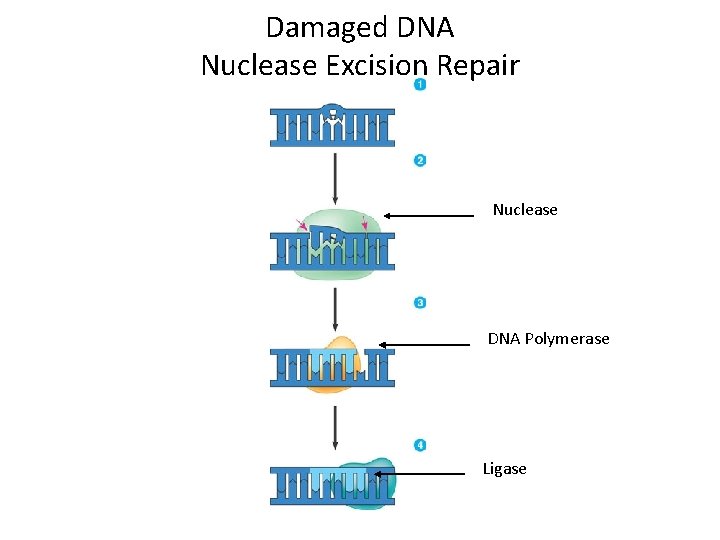
Damaged DNA Nuclease Excision Repair Nuclease DNA Polymerase Ligase

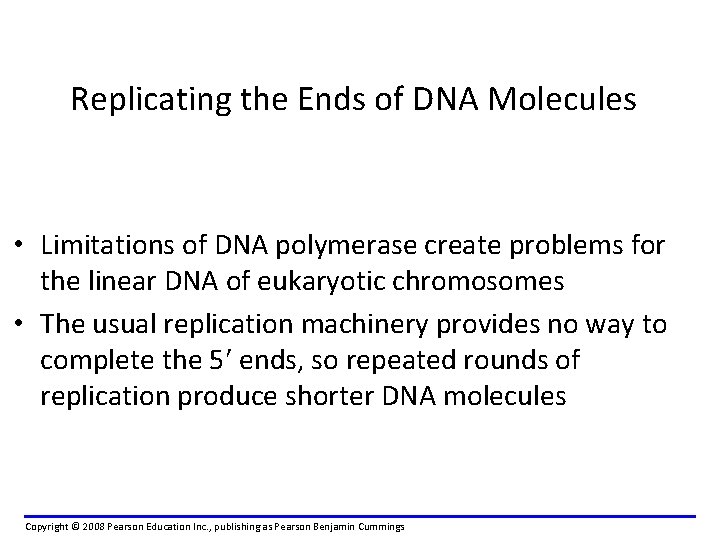
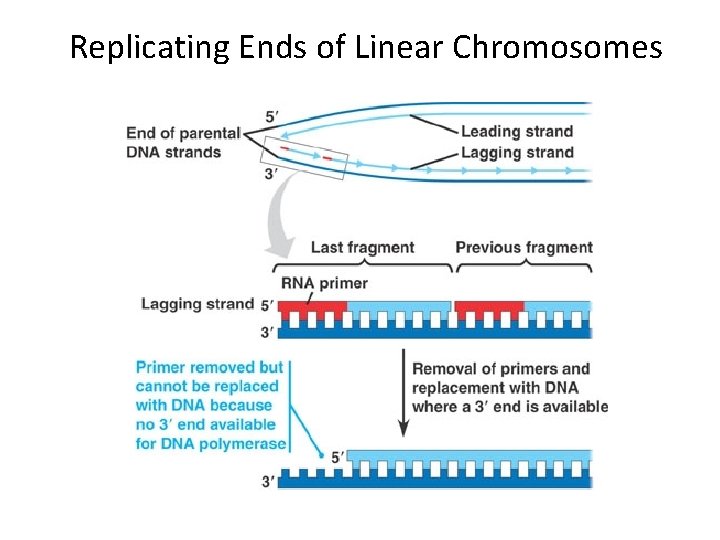
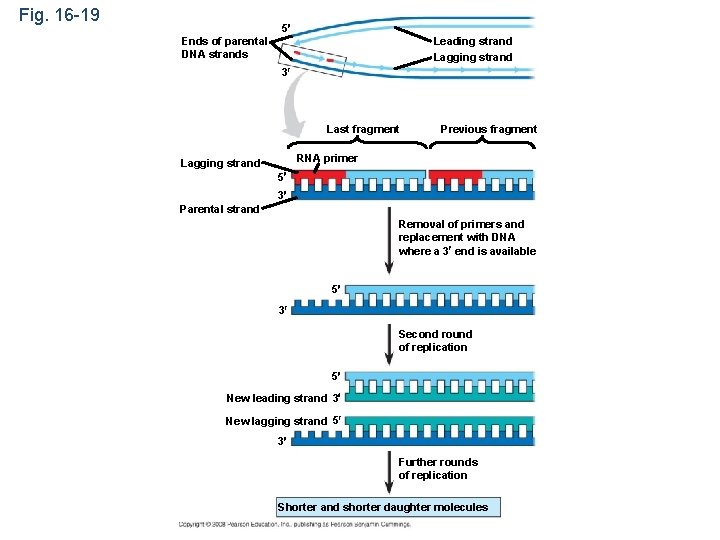
Replicating the Ends of DNA Molecules • Limitations of DNA polymerase create problems for the linear DNA of eukaryotic chromosomes • The usual replication machinery provides no way to complete the 5 ends, so repeated rounds of replication produce shorter DNA molecules Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Replicating Ends of Linear Chromosomes

Fig. 16 -19 5 Ends of parental DNA strands Leading strand Lagging strand 3 Last fragment Previous fragment RNA primer Lagging strand 5 3 Parental strand Removal of primers and replacement with DNA where a 3 end is available 5 3 Second round of replication 5 New leading strand 3 New lagging strand 5 3 Further rounds of replication Shorter and shorter daughter molecules

Fig. 16 -20 1 µm

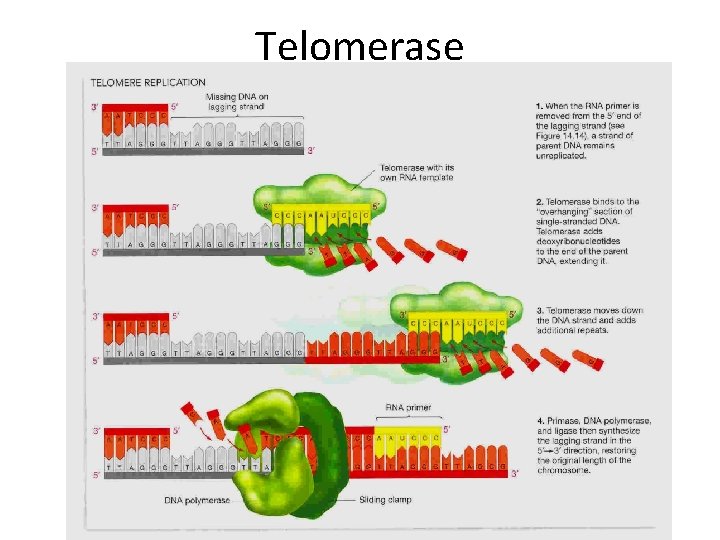
• If chromosomes of germ cells became shorter in every cell cycle, essential genes would eventually be missing from the gametes they produce • An enzyme called telomerase catalyzes the lengthening of telomeres in germ cells Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Telomerase

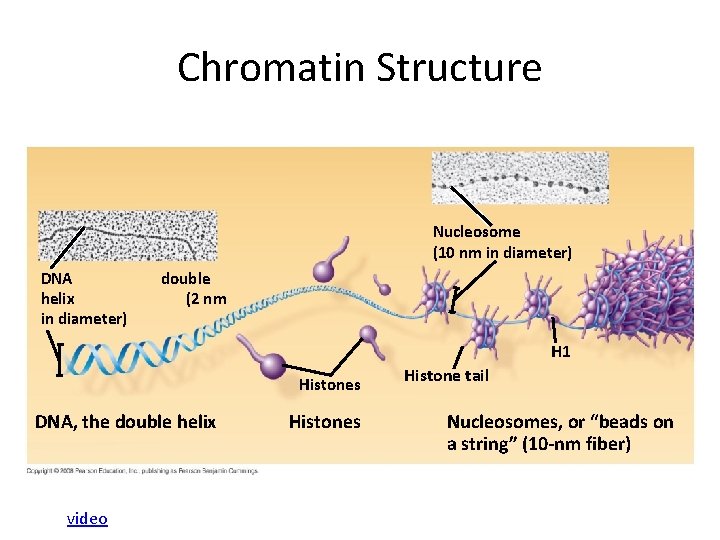
Chromatin Structure Nucleosome (10 nm in diameter) DNA helix in diameter) double (2 nm H 1 Histones DNA, the double helix video Histones Histone tail Nucleosomes, or “beads on a string” (10 -nm fiber)

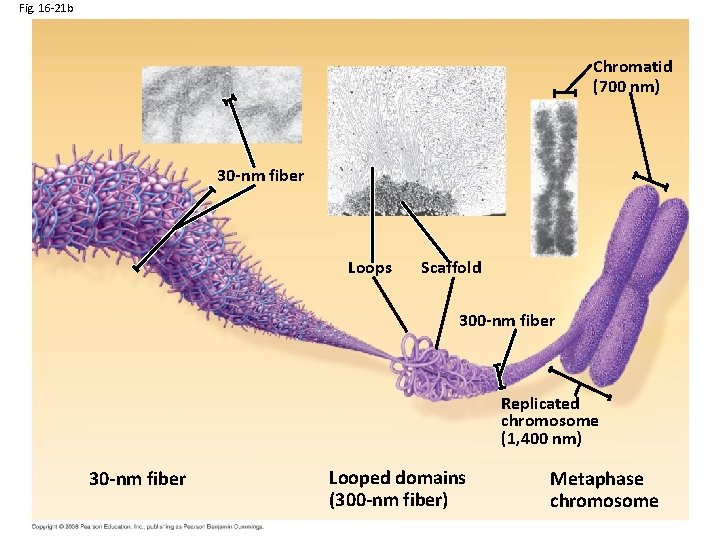
Fig. 16 -21 b Chromatid (700 nm) 30 -nm fiber Loops Scaffold 300 -nm fiber Replicated chromosome (1, 400 nm) 30 -nm fiber Looped domains (300 -nm fiber) Metaphase chromosome

30 nm chromatin fiber 1. Held together by histone tails interacting with neighboring nucleosomes 2. Inhibits transcription 3. Allows DNA replication

Gene Expression • • Beadle and Tatum Exp Transcription Translation Roles of RNA

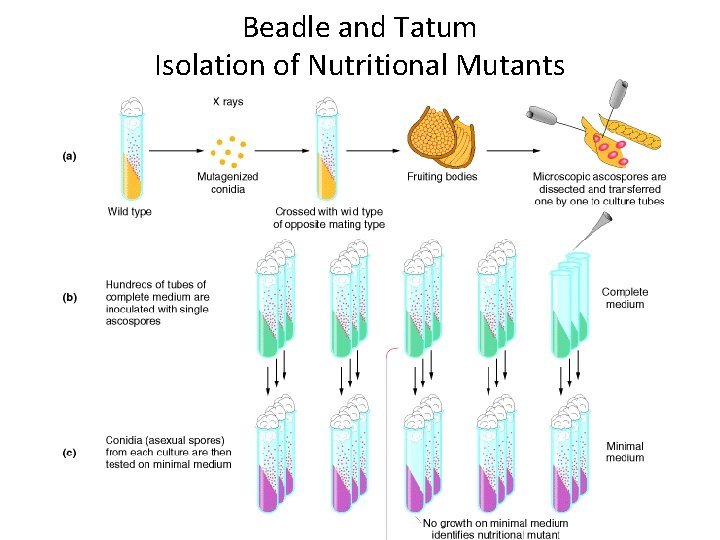
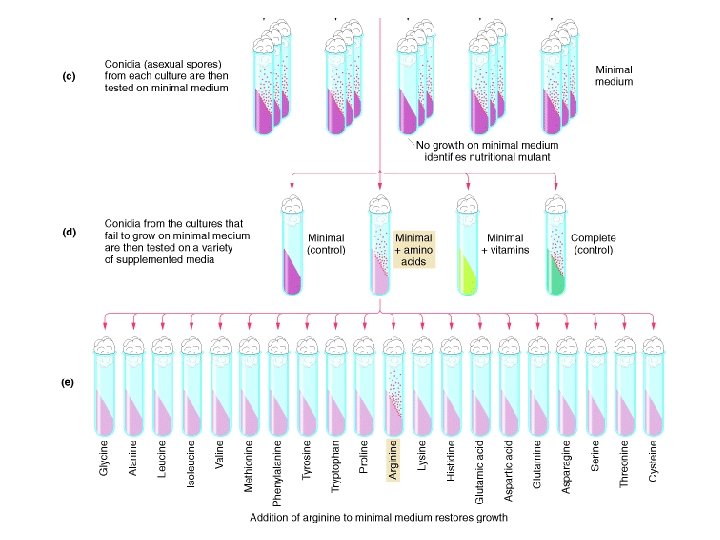
Beadle and Tatum Isolation of Nutritional Mutants


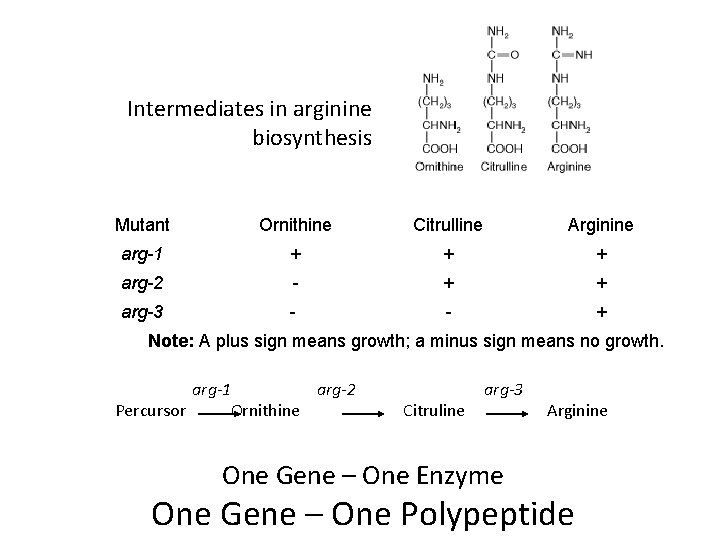
Intermediates in arginine biosynthesis Mutant Ornithine Citrulline Arginine arg-1 + + + arg-2 - + + arg-3 - - + Note: A plus sign means growth; a minus sign means no growth. arg-1 arg-2 Percursor Ornithine Citruline arg-3 One Gene – One Enzyme Arginine One Gene – One Polypeptide

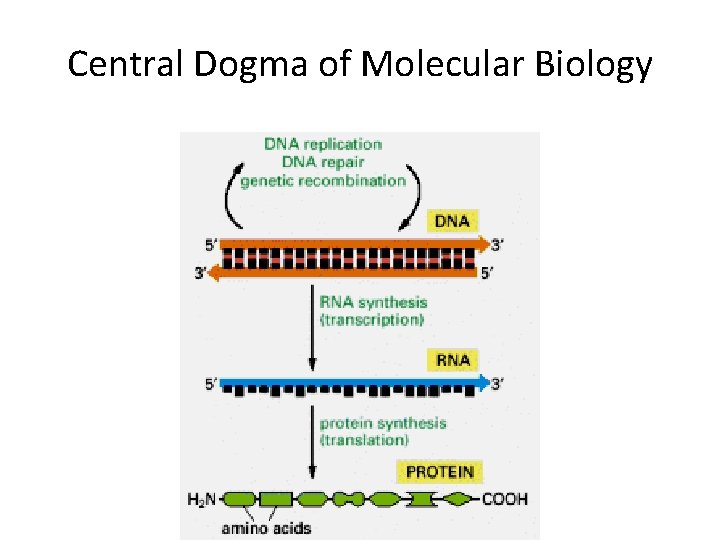
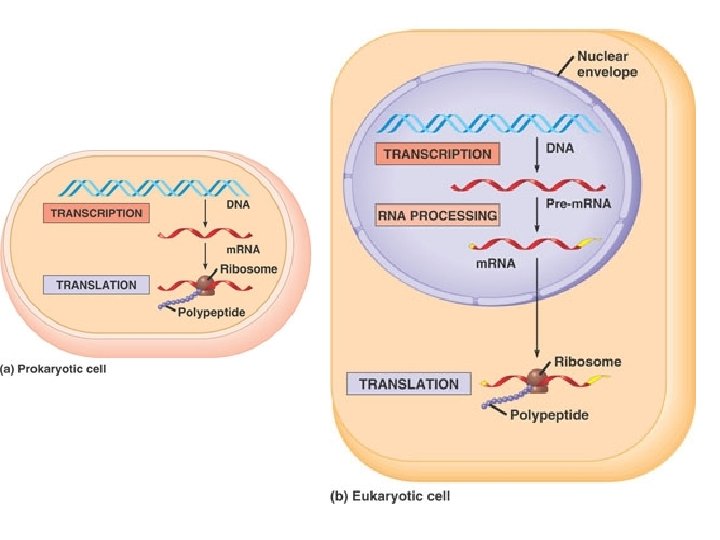
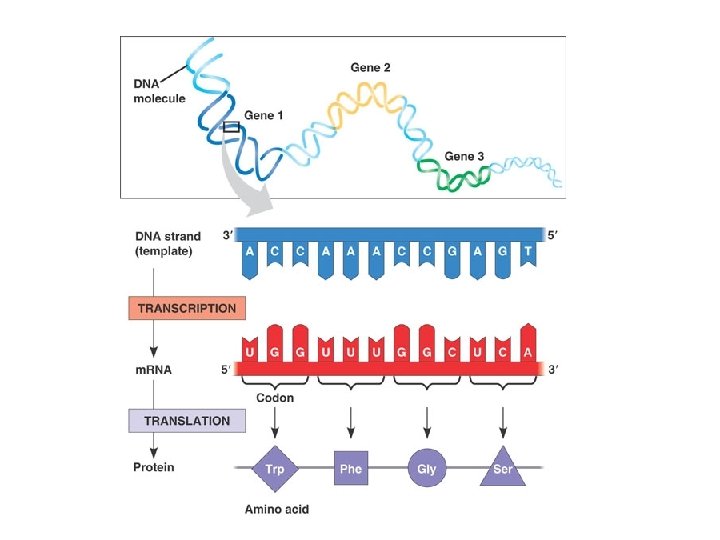
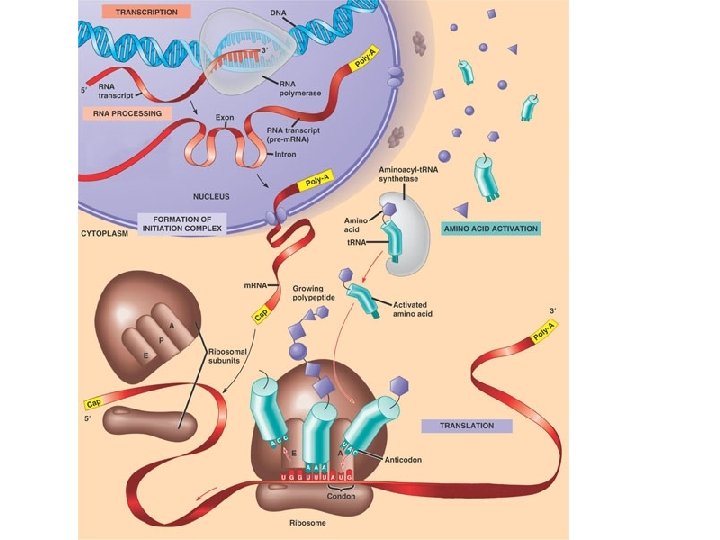
Central Dogma of Molecular Biology



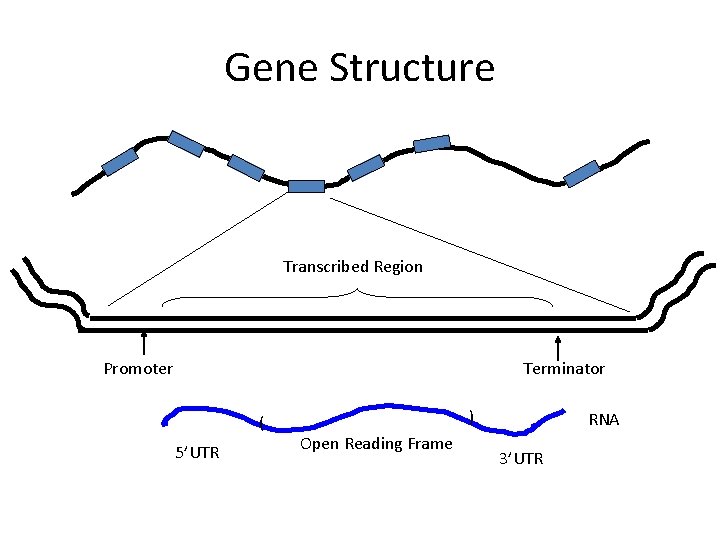
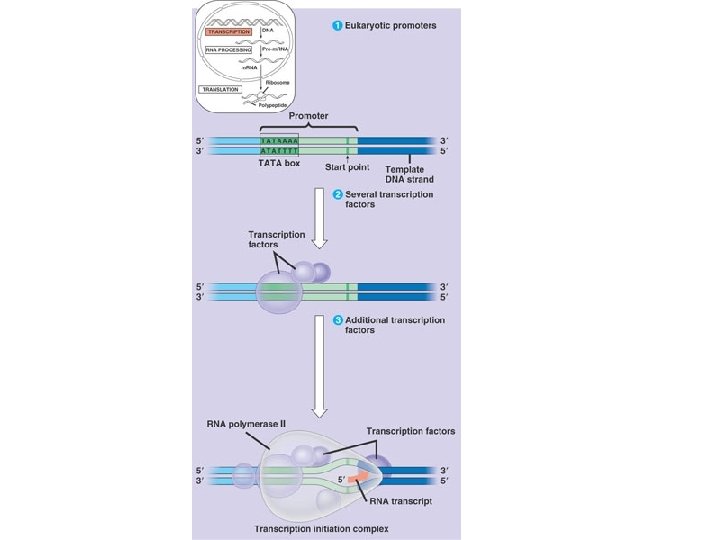
Gene Structure Transcribed Region Promoter Terminator ( 5’UTR ) Open Reading Frame RNA 3’UTR


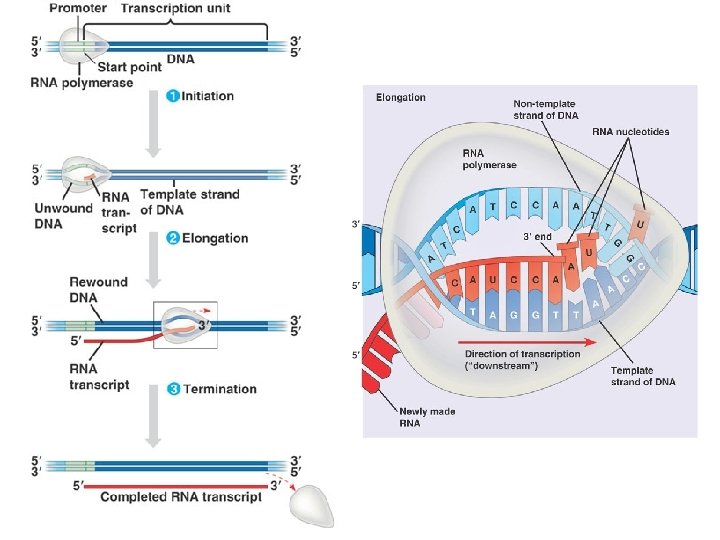
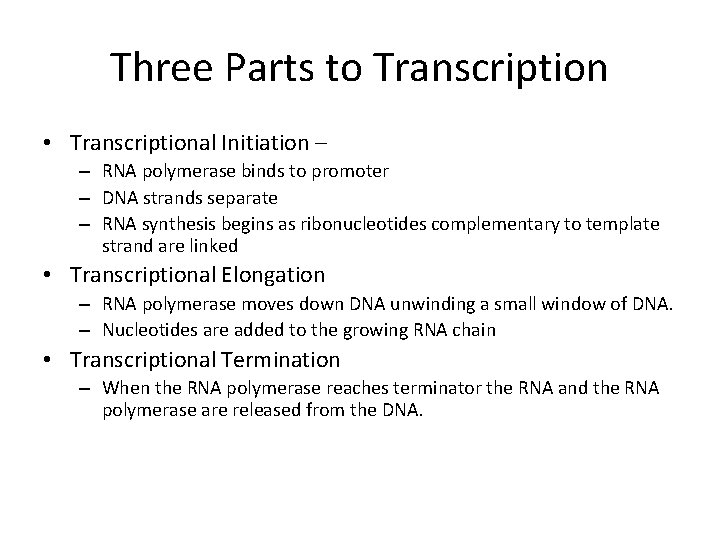
Three Parts to Transcription • Transcriptional Initiation – – RNA polymerase binds to promoter – DNA strands separate – RNA synthesis begins as ribonucleotides complementary to template strand are linked • Transcriptional Elongation – RNA polymerase moves down DNA unwinding a small window of DNA. – Nucleotides are added to the growing RNA chain • Transcriptional Termination – When the RNA polymerase reaches terminator the RNA and the RNA polymerase are released from the DNA.


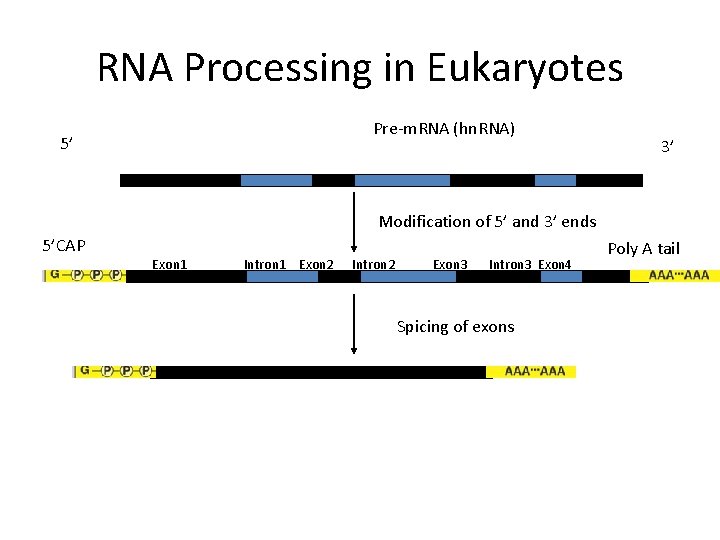
RNA Processing in Eukaryotes Pre-m. RNA (hn. RNA) 5’ 3’ Modification of 5’ and 3’ ends 5’CAP Exon 1 Intron 1 Exon 2 Intron 2 Exon 3 Intron 3 Exon 4 Spicing of exons Poly A tail


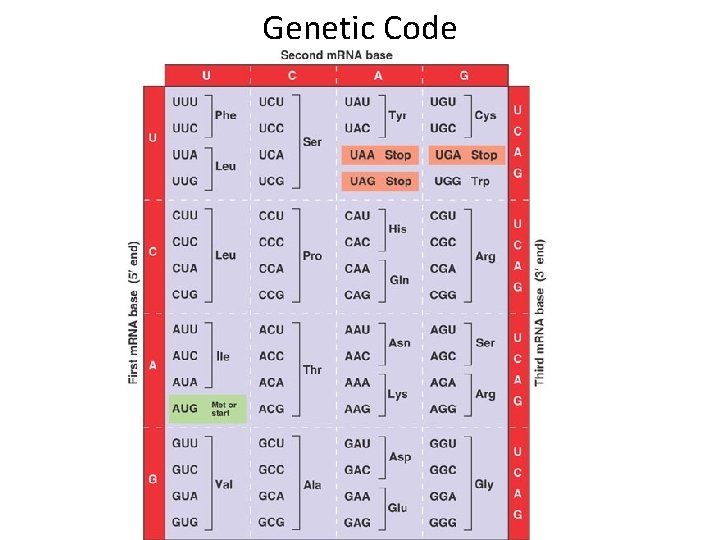
Genetic Code

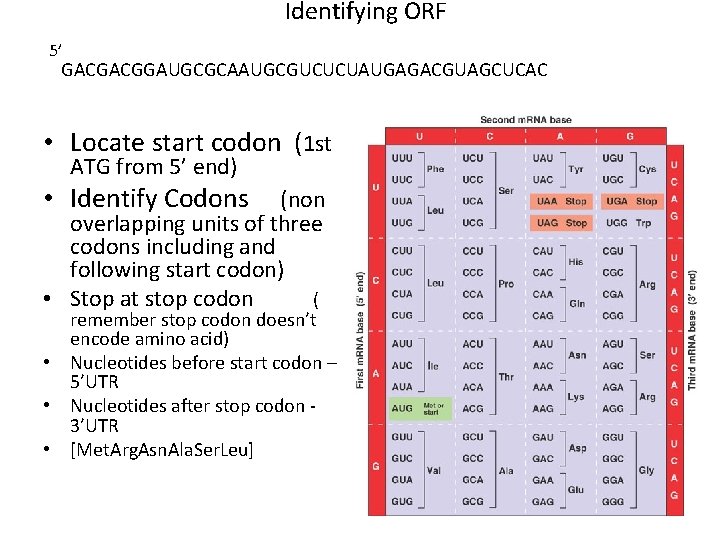
Identifying ORF 5’ GACGACGGAUGCGCAAUGCGUCUCUAUGAGACGUAGCUCAC • Locate start codon (1 st ATG from 5’ end) • Identify Codons (non overlapping units of three codons including and following start codon) • Stop at stop codon ( remember stop codon doesn’t encode amino acid) • Nucleotides before start codon – 5’UTR • Nucleotides after stop codon 3’UTR • [Met. Arg. Asn. Ala. Ser. Leu]

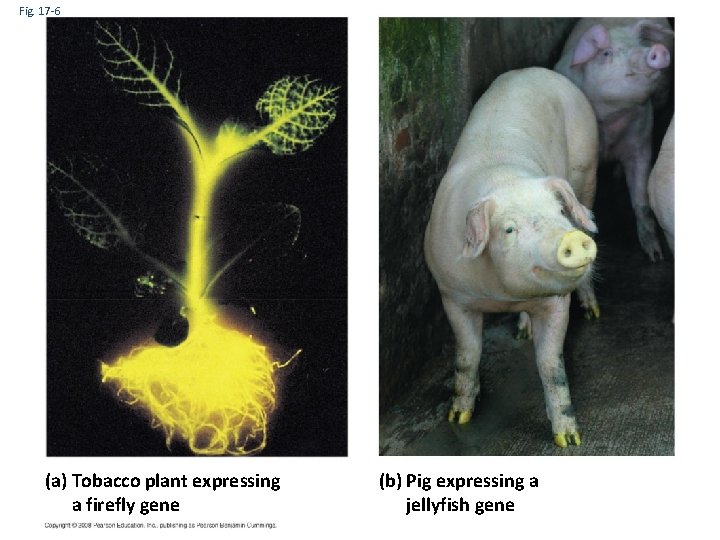
Fig. 17 -6 (a) Tobacco plant expressing a firefly gene (b) Pig expressing a jellyfish gene

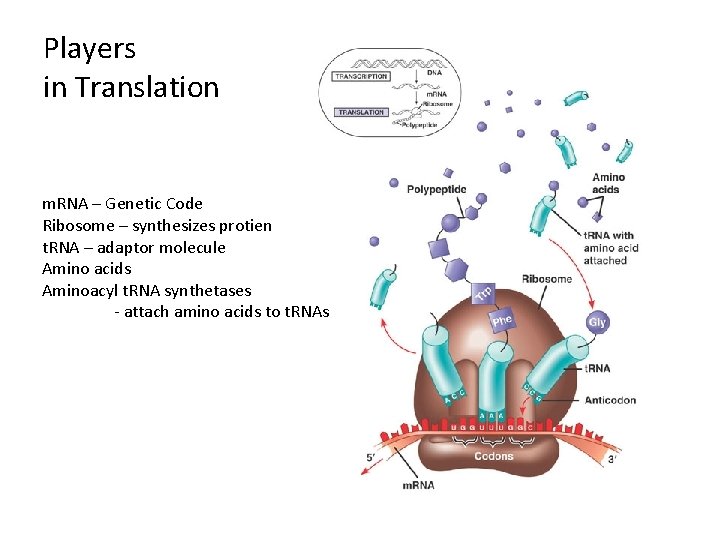
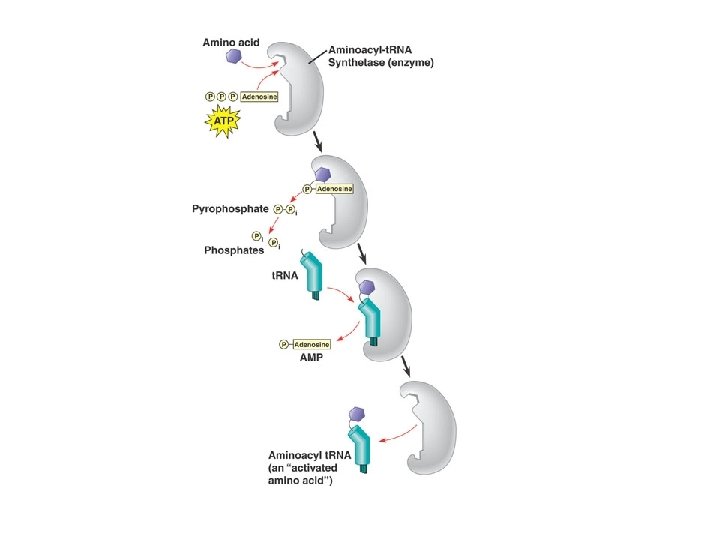
Players in Translation m. RNA – Genetic Code Ribosome – synthesizes protien t. RNA – adaptor molecule Amino acids Aminoacyl t. RNA synthetases - attach amino acids to t. RNAs

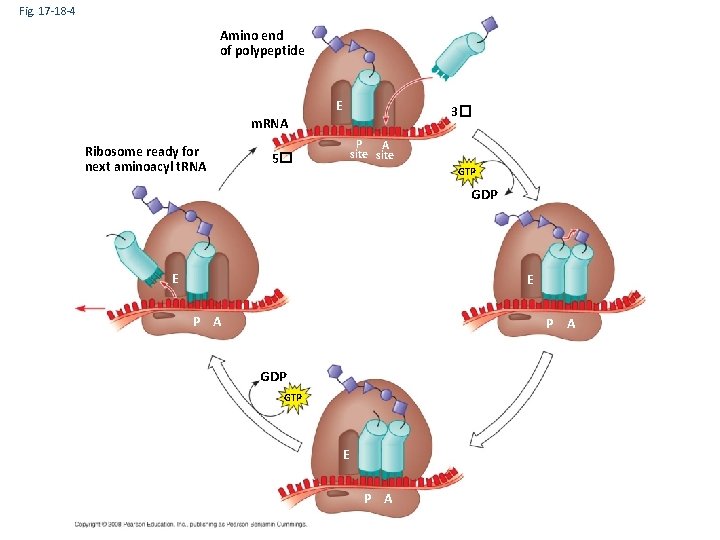
Fig. 17 -18 -4 Amino end of polypeptide E 3� m. RNA Ribosome ready for next aminoacyl t. RNA P A site 5� GTP GDP E E P A GDP GTP E P A

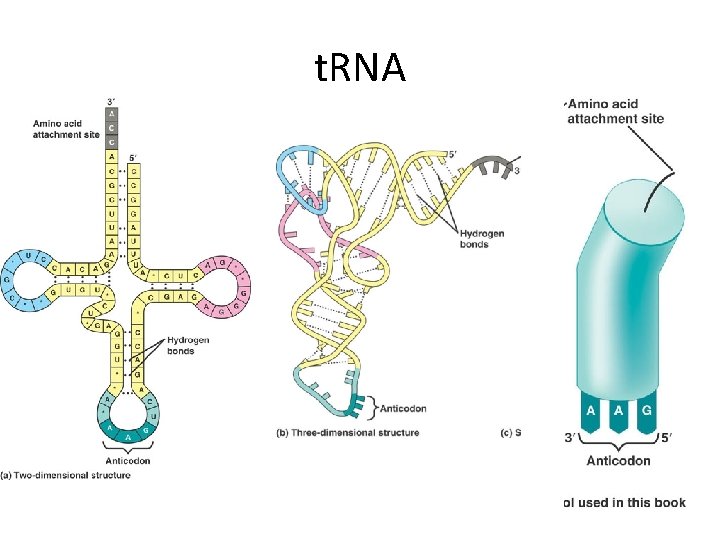
t. RNA


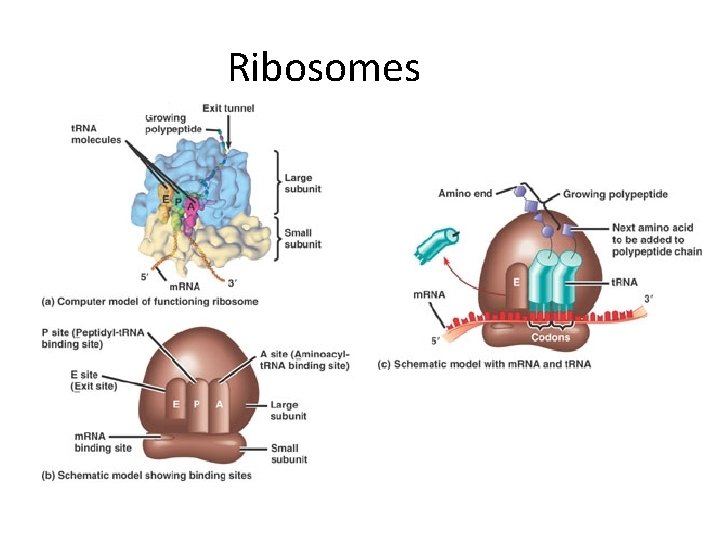
Ribosomes

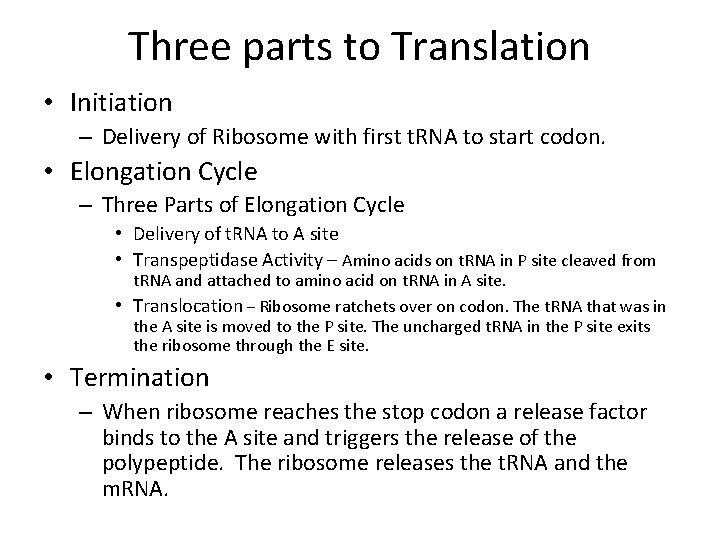
Three parts to Translation • Initiation – Delivery of Ribosome with first t. RNA to start codon. • Elongation Cycle – Three Parts of Elongation Cycle • Delivery of t. RNA to A site • Transpeptidase Activity – Amino acids on t. RNA in P site cleaved from t. RNA and attached to amino acid on t. RNA in A site. • Translocation – Ribosome ratchets over on codon. The t. RNA that was in the A site is moved to the P site. The uncharged t. RNA in the P site exits the ribosome through the E site. • Termination – When ribosome reaches the stop codon a release factor binds to the A site and triggers the release of the polypeptide. The ribosome releases the t. RNA and the m. RNA.

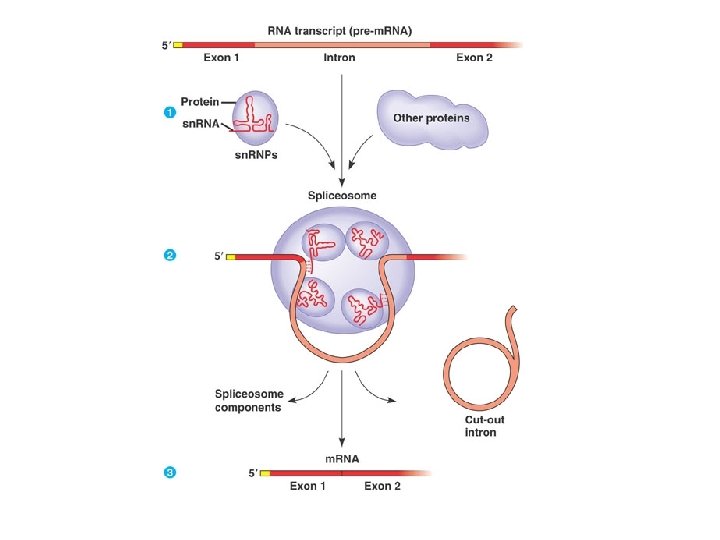
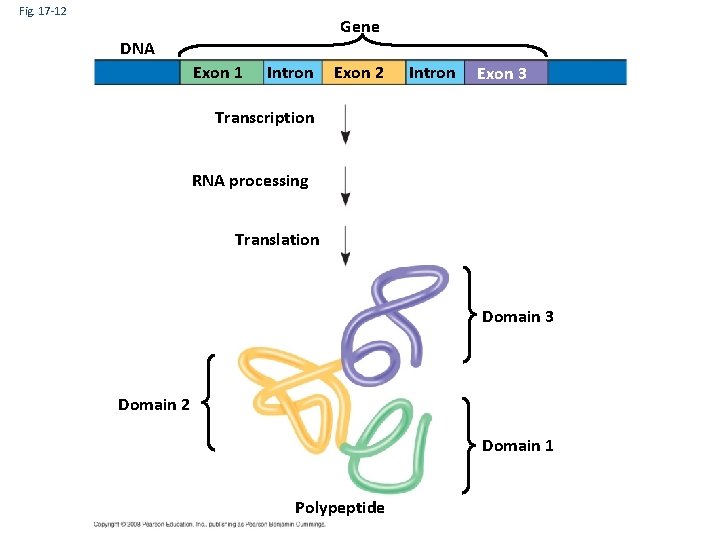
The Functional and Evolutionary Importance of Introns • Some genes can encode more than one kind of polypeptide, depending on which segments are treated as exons during RNA splicing • Such variations are called alternative RNA splicing • Because of alternative splicing, the number of different proteins an organism can produce is much greater than its number of genes Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Fig. 17 -12 Gene DNA Exon 1 Intron Exon 2 Intron Exon 3 Transcription RNA processing Translation Domain 3 Domain 2 Domain 1 Polypeptide

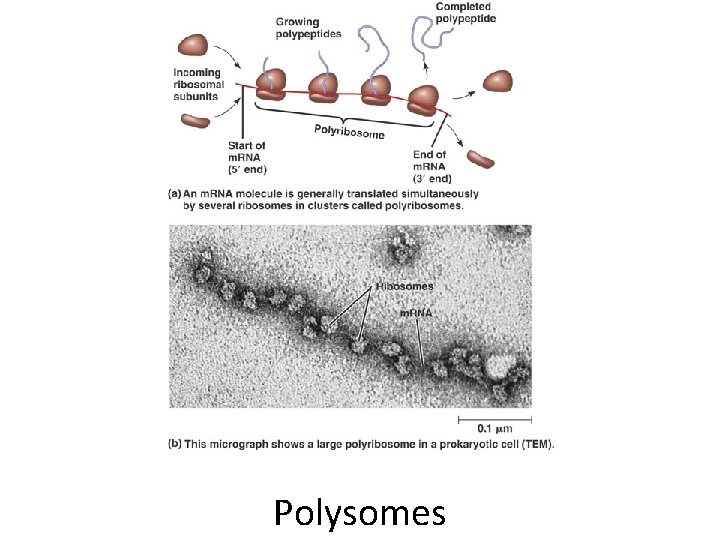
Polysomes

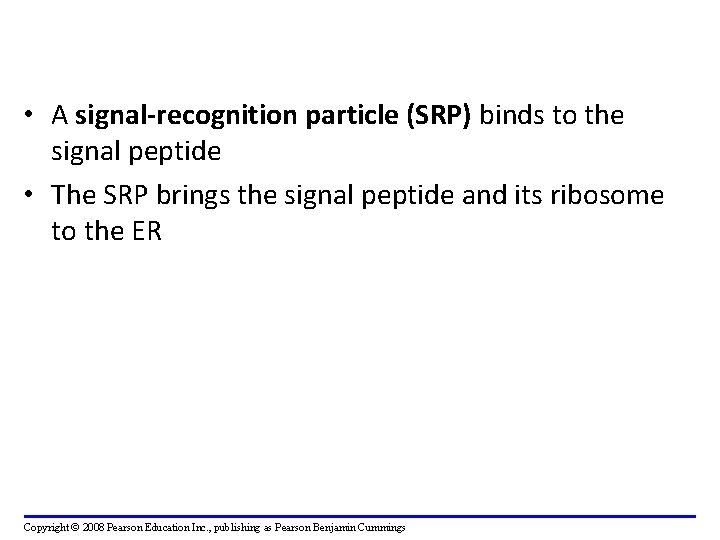
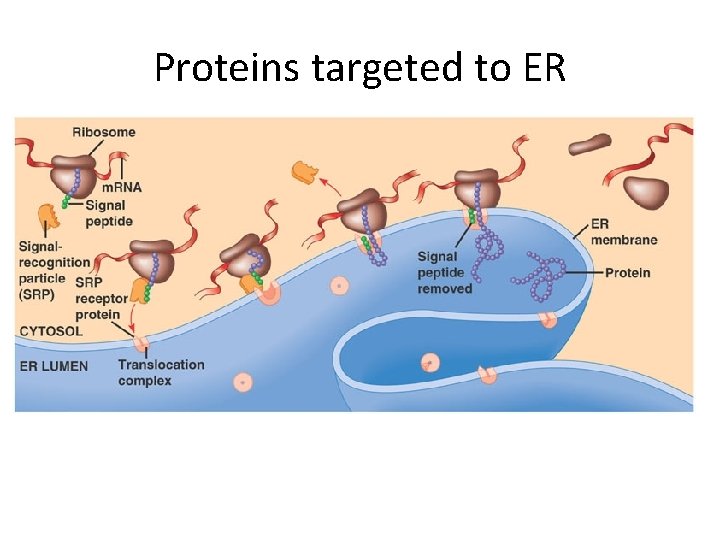
• Polypeptide synthesis always begins in the cytosol • Synthesis finishes in the cytosol unless the polypeptide signals the ribosome to attach to the ER • Polypeptides destined for the ER or for secretion are marked by a signal peptide Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

• A signal-recognition particle (SRP) binds to the signal peptide • The SRP brings the signal peptide and its ribosome to the ER Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Proteins targeted to ER

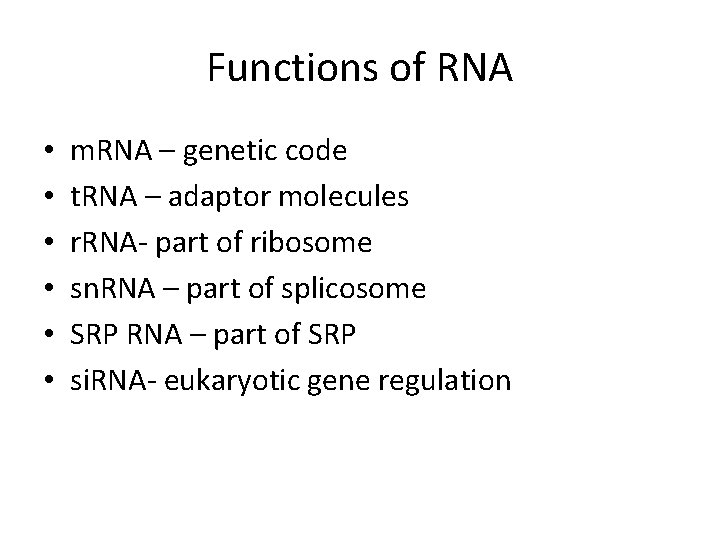
Functions of RNA • • • m. RNA – genetic code t. RNA – adaptor molecules r. RNA- part of ribosome sn. RNA – part of splicosome SRP RNA – part of SRP si. RNA- eukaryotic gene regulation

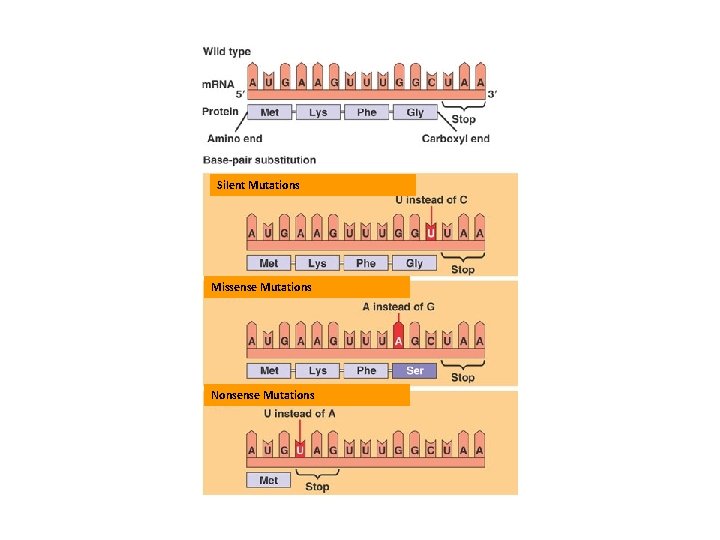
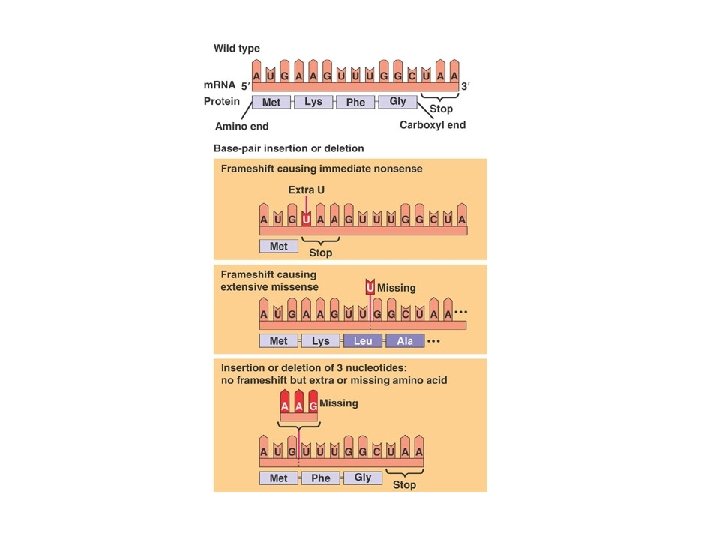
Silent Mutations Missense Mutations Nonsense Mutations

