Structural Effects of Cytidine 2 Ribose Modifications as
Structural Effects of Cytidine 2’ Ribose Modifications as Determined by IRMPD Action Spectroscopy Lucas Hamlow†, Chenchen He†, Lin Fan†, Ranran Wu†, Bo Yang†, Giel Berden‡, Jos Oomens‡ and M. T. Rodgers† †Department of Chemistry, Wayne State University, Detroit, MI ‡ Institute for Molecules and Materials, Radboud University Nijmegen, FELIX Facility, The Netherlands ISMS June 22, 2015

Modified Nucleosides • Modified nucleosides play an important role in the function of a variety of RNAs • 4 main type of modifications: – – Uridine pseudouridine Nucleobase modifications 2’ ribose hydroxyl methylation Multiple modifications • Nucleoside analogs are used to treat a variety of diseases – – AIDS/HIV Hepatitis B Cancer Etc… C. B. Lauter, E. J. Bailey, A. M. Lerner Antimicrob. Agents Chemother. 598 (1974) P. Liu, A. Sharon, C. K. Chu J Fluor Chem 129(9) 743 (2008)
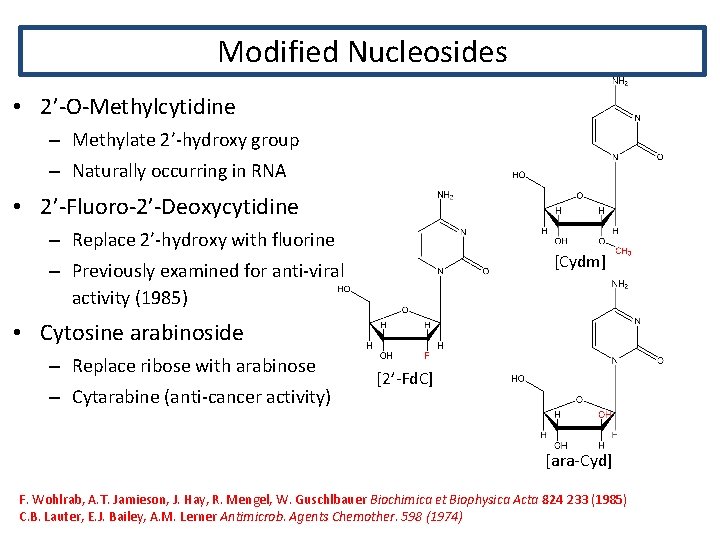
Modified Nucleosides • 2’-O-Methylcytidine – Methylate 2’-hydroxy group – Naturally occurring in RNA • 2’-Fluoro-2’-Deoxycytidine – Replace 2’-hydroxy with fluorine [Cydm] – Previously examined for anti-viral activity (1985) • Cytosine arabinoside – Replace ribose with arabinose – Cytarabine (anti-cancer activity) [2’-Fd. C] [ara-Cyd] F. Wohlrab, A. T. Jamieson, J. Hay, R. Mengel, W. Guschlbauer Biochimica et Biophysica Acta 824 233 (1985) C. B. Lauter, E. J. Bailey, A. M. Lerner Antimicrob. Agents Chemother. 598 (1974)
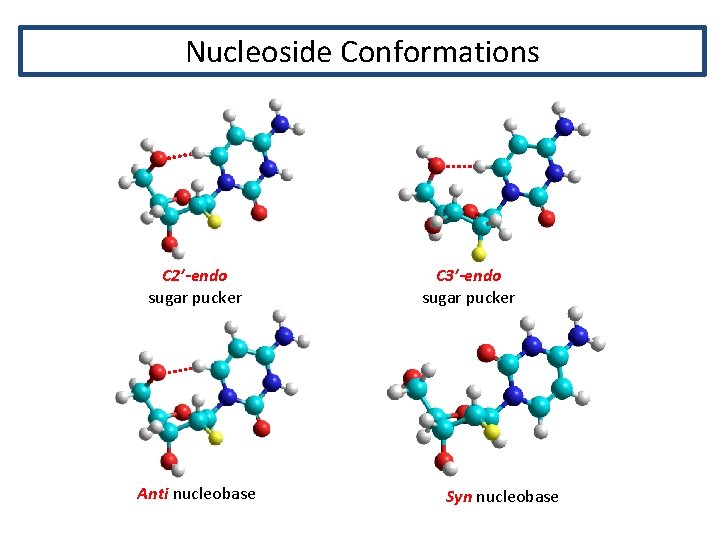
Nucleoside Conformations C 2’-endo sugar pucker Anti nucleobase C 3’-endo sugar pucker Syn nucleobase
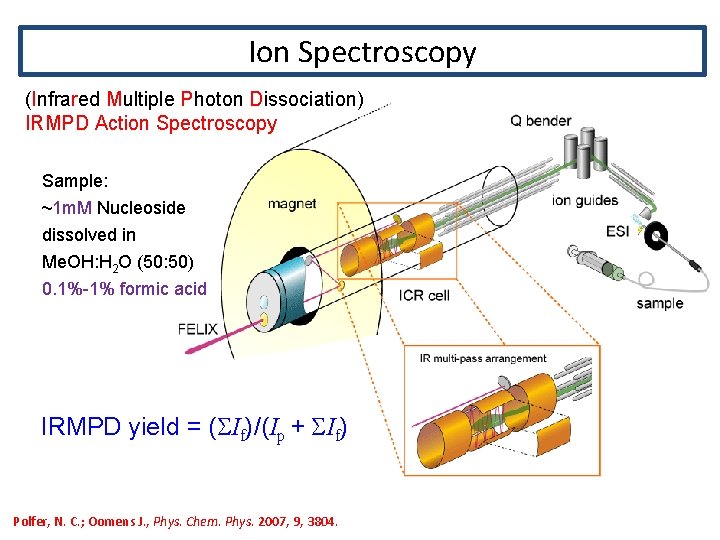
Ion Spectroscopy (Infrared Multiple Photon Dissociation) IRMPD Action Spectroscopy Sample: ~1 m. M Nucleoside dissolved in Me. OH: H 2 O (50: 50) 0. 1%-1% formic acid IRMPD yield = (SIf)/(Ip + SIf) Polfer, N. C. ; Oomens J. , Phys. Chem. Phys. 2007, 9, 3804.
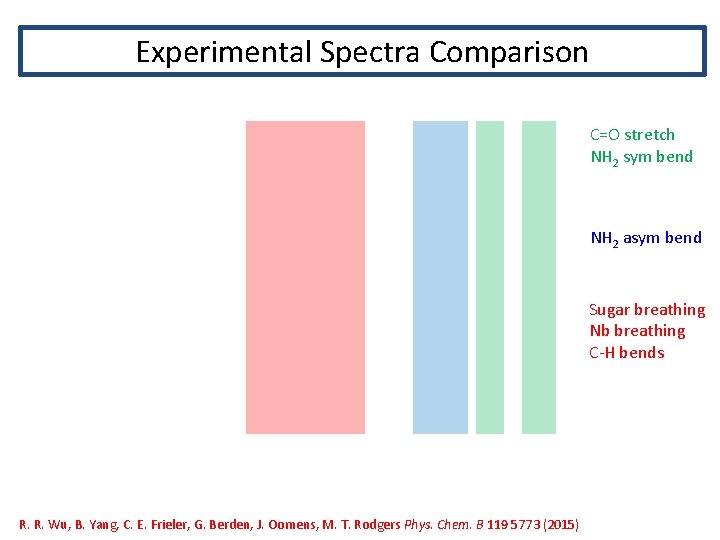
Experimental Spectra Comparison C=O stretch NH 2 sym bend NH 2 asym bend Sugar breathing Nb breathing C-H bends R. R. Wu, B. Yang, C. E. Frieler, G. Berden, J. Oomens, M. T. Rodgers Phys. Chem. B 119 5773 (2015)
Structure Calculations • Candidate structure selection – Simulated annealing using Amber 3 force field generates 300 candidate structures – Lowest 30 in energy are chosen for DFT calculations • Electronic structure calculations – Geometry optimizations and frequency analyses done with B 3 LYP/6 -311+G(d, p) – Energy calculations done with B 3 LYP/6 -311+G(2 d, 2 p) and MP 2/6 -311+G(2 d, 2 p) • Convoluted theoretical spectra – Gaussian peak shapes with 20 cm-1 FWHM – Single scaling factor applied to best match experimental spectra
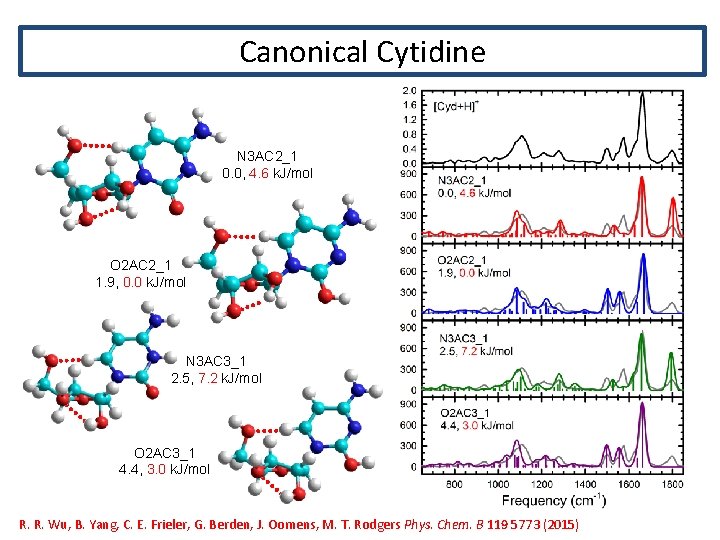
Canonical Cytidine N 3 AC 2_1 0. 0, 4. 6 k. J/mol O 2 AC 2_1 1. 9, 0. 0 k. J/mol N 3 AC 3_1 2. 5, 7. 2 k. J/mol O 2 AC 3_1 4. 4, 3. 0 k. J/mol R. R. Wu, B. Yang, C. E. Frieler, G. Berden, J. Oomens, M. T. Rodgers Phys. Chem. B 119 5773 (2015)
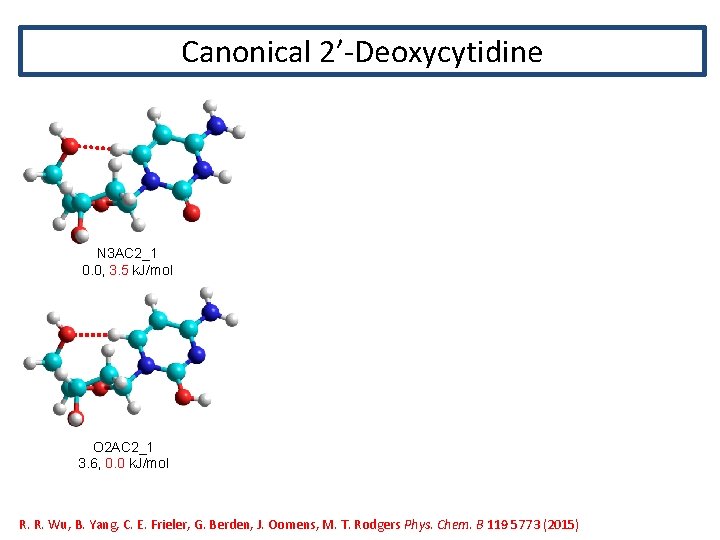
Canonical 2’-Deoxycytidine N 3 AC 2_1 0. 0, 3. 5 k. J/mol O 2 AC 2_1 3. 6, 0. 0 k. J/mol R. R. Wu, B. Yang, C. E. Frieler, G. Berden, J. Oomens, M. T. Rodgers Phys. Chem. B 119 5773 (2015)
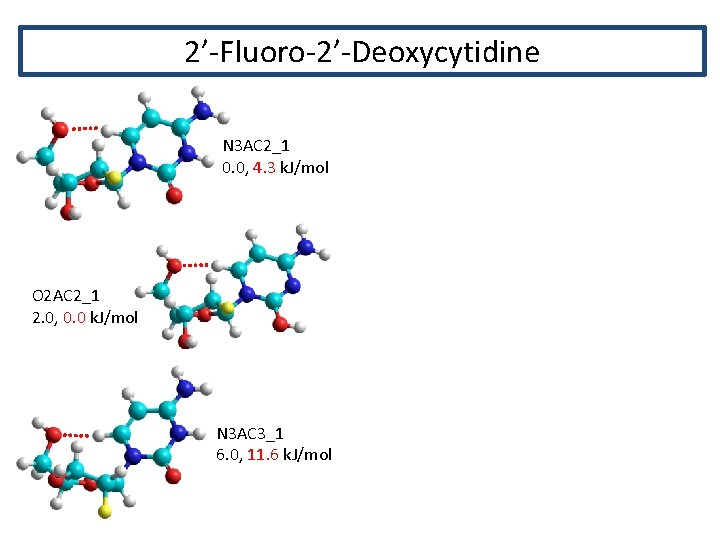
2’-Fluoro-2’-Deoxycytidine N 3 AC 2_1 0. 0, 4. 3 k. J/mol O 2 AC 2_1 2. 0, 0. 0 k. J/mol N 3 AC 3_1 6. 0, 11. 6 k. J/mol

Cytosine Arabinoside N 3 AC 2_1 0. 0, 1. 5 k. J/mol N 3 AC 2_2 3. 3, 4. 8 k. J/mol O 2 AC 2_1 5. 4, 0. 0 k. J/mol A. Filippi et al. Int. Journal of Mass Spectrometry 354 54 (2013)

2’-O-Methylcytidine N 3 AC 3_1 0. 0, 0. 5 k. J/mol N 3 AC 2_1 6. 0, 6. 0 k. J/mol O 2 AC 2_1 6. 7, 0. 0 k. J/mol
![Stability of N 3 and O 2 Protonation • [Cyd+H]+ and [d. Cyd+H]+ – Stability of N 3 and O 2 Protonation • [Cyd+H]+ and [d. Cyd+H]+ –](http://slidetodoc.com/presentation_image/8fe5e8254b4ab3d2da2740fda89c9603/image-13.jpg)
Stability of N 3 and O 2 Protonation • [Cyd+H]+ and [d. Cyd+H]+ – Roughly equal populations of N 3 and O 2 protonation predicted by MP 2 and B 3 LYP • [2’-Fd. C+H]+ – B 3 LYP overestimates the stability of N 3 protonation while the spectra would suggest roughly equal populations • [ara-Cyd+H]+ – B 3 LYP overestimates the stability of N 3 protonation but does predict a small preference for N 3 which the spectra would suggest • [Cydm+H]+ – MP 2 predicts O 2 protonation to be very slightly preferred which matches best with the spectra R. R. Wu, B. Yang, C. E. Frieler, G. Berden, J. Oomens, M. T. Rodgers Phys. Chem. B 119 5773 (2015)
![Structure Comparison • [2’-Fd. C+H]+ – Little structural change from [d. Cyd+H]+ – Must Structure Comparison • [2’-Fd. C+H]+ – Little structural change from [d. Cyd+H]+ – Must](http://slidetodoc.com/presentation_image/8fe5e8254b4ab3d2da2740fda89c9603/image-14.jpg)
Structure Comparison • [2’-Fd. C+H]+ – Little structural change from [d. Cyd+H]+ – Must have an electronic effect [Cyd+H]+ • [ara-Cyd+H]+ – Subtle shift in glycosidic bond angle – Will likely have a structural effect [2’Fd. Cyd+H]+ [d. Cyd+H]+ • [Cydm+H]+ – Slight preference for O 2 with similar structure to the above over a C 3’-endo N 3 structure [ara-Cyd+H]+ [Cydm+H]+ R. R. Wu, B. Yang, C. E. Frieler, G. Berden, J. Oomens, M. T. Rodgers Phys. Chem. B 119 5773 (2015)

Acknowledgements • • • Rodgers group members Dr. M. T. Rodgers Dr. Jos Oomens, Dr. Giel Berden FELIX support staff WSU C&IT CLIO Facility Department of Chemistry FELIX Facility
- Slides: 15