String Manipulations Web Scraping MultilevelModeling Population Genetics in

String Manipulations, Web. Scraping, Multilevel-Modeling, Population Genetics in R Session 19, 11/5/2018 Nick Brazeau

AKA Nick’s potpourri

Outline • String manipulations • Web-scraping • Multilevel Modeling • Stop Here • Population Genetics/Genomics in R

String Manipulations in R stringr

String Manipulations • Remember in R that all strings are of type “char” • We can use the stringr package to manipulate, parse, detect, etc. these strings • Looking forward: • Much of population genetic haplotypic analyses is just string manipulations

My favorite stringr commands • str_split (and str_split_fixed) • str_extract • str_detect • Many, many more!

Plan • Go to Rmarkdown for some examples • Vignette on your own (5 -10 minutes)

stringr cheat sheet
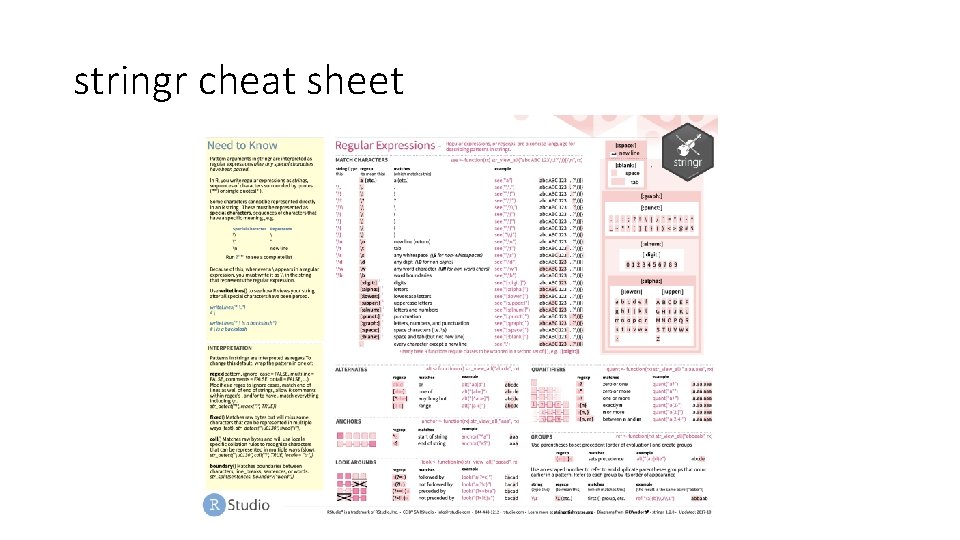
stringr cheat sheet

Web Scraping in R

Web Scraping • The process of extracting data from the web (i. e. a website, etc. ) • Take html code and convert it into a data format we can use • With great power, comes great responsibility (careful with serverload) • Some sites do not “allow” scraping. Laws behind this are evolving… • ^^ More of an issue for company espionage, not querying servers that are clearly meant for public resources (i. e. ncbi, plasmodb…)
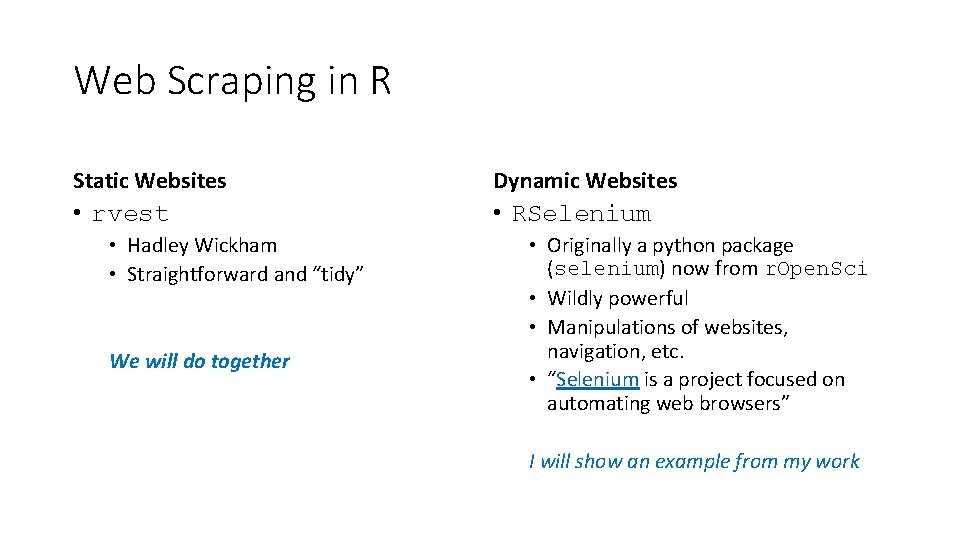
Web Scraping in R Static Websites Dynamic Websites • rvest • RSelenium • Hadley Wickham • Straightforward and “tidy” We will do together • Originally a python package (selenium) now from r. Open. Sci • Wildly powerful • Manipulations of websites, navigation, etc. • “Selenium is a project focused on automating web browsers” I will show an example from my work

Plan 1 • RSelenium example from my work • rvest tutorial from Hadley Wickham • Advised to download the “Selector. Gadget” tool • Can also open up html code in google chrome using inspect • Regardless, read the vignette("selectorgadget") from the rvest package to get a sense of how to select components of a website based on their css type 5 -10 Minute Breakout to do this Vignette and Tutorial, Lego-Movie

Plan 2 Today’s Main Exercise 1. “Scrape” the average income of each county from Wikipedia. 2. Merge this information into our births dataset 3. Then we will simulate an income covariate for each individual in our dataset based on the average with some ”noise” added • Means we are creating a variable that violates the assumption of independence in regression modeling = PROBLEM (NEXT) 4. Perform multilevel (a solution)

Multilevel Modeling in R

Acknowledgements • Our. Coding. Club • OJ Watson • ME 3
Multilevel Modeling Use Case • Non-independence among covariates • Repeated measurements (or the same patient/person/individual) • Sampling from a classroom, village, county, etc. • Clusters differ systematically • This means we violates our independence assumption in regression Funny/Helpful Article: Keep Calm and Learn Multilevel Logistic Modeling: A Simplified Three-Step Procedure Using Stata, R, Mplus, and SPSS

Brief Notes • Essentially, we can fit a different slope and/or intercept to each cluster (intracluster— “random” effect) while accounting for an overall “fixed” effect • This means, we are now trying to determine the [risk, odds, etc] of someone with treatment exposure, i, that is part of cluster, j. Funny/Helpful Article: Keep Calm and Learn Multilevel Logistic Modeling: A Simplified Three-Step Procedure Using Stata, R, Mplus, and SPSS
Brief Notes Screenshot from Our. Coding. Club

Brief Notes • For more in depth tutorials, check out: • Our. Coding. Club • OJ Watson’s tutorial • https: //www. r-bloggers. com/multilevel-modeling-of-educational-data-using-r-part-1/ • https: //www. jaredknowles. com/journal/2013/11/25/getting-started-with-mixed-effect-models-in-r • Many, many more

Plan • Rmarkdown example
Intro to Pop Gen (for those who are interested)

Outline • String manipulations • Web-scraping • Multilevel Modeling • Stop Here • Population Genetics/Genomics in R

Population Genetics & Genomics in R
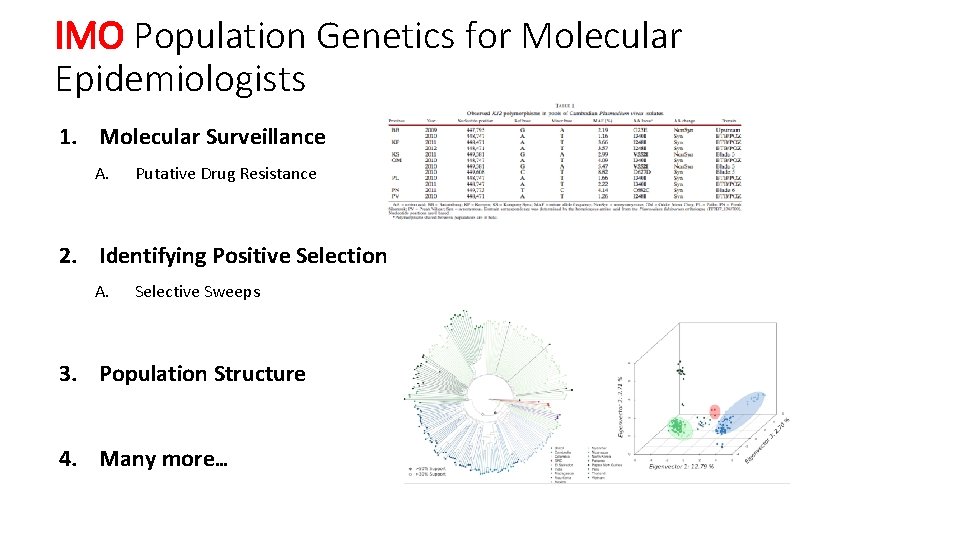
IMO Population Genetics for Molecular Epidemiologists 1. Molecular Surveillance A. Putative Drug Resistance 2. Identifying Positive Selection A. Selective Sweeps 3. Population Structure 4. Many more…
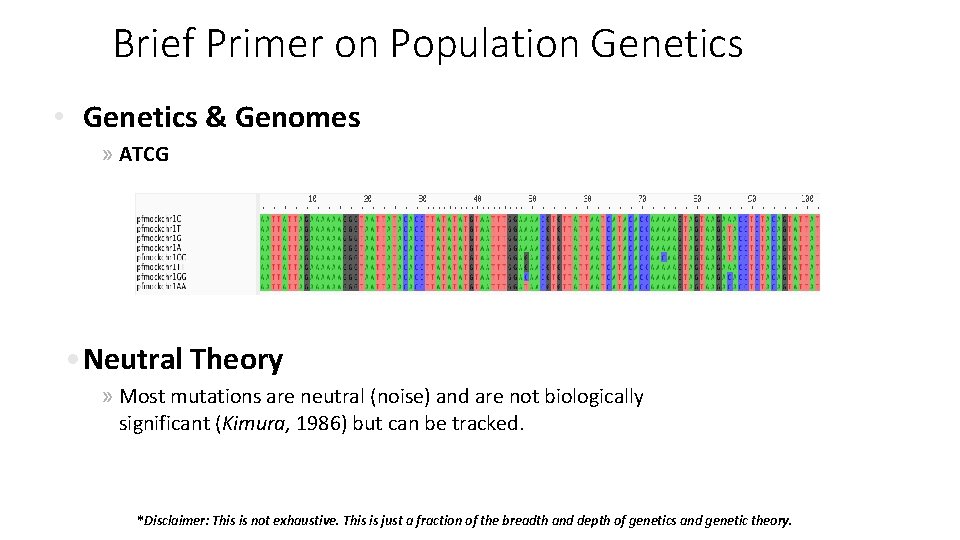
Brief Primer on Population Genetics • Genetics & Genomes » ATCG • Neutral Theory » Most mutations are neutral (noise) and are not biologically significant (Kimura, 1986) but can be tracked. *Disclaimer: This is not exhaustive. This is just a fraction of the breadth and depth of genetics and genetic theory.
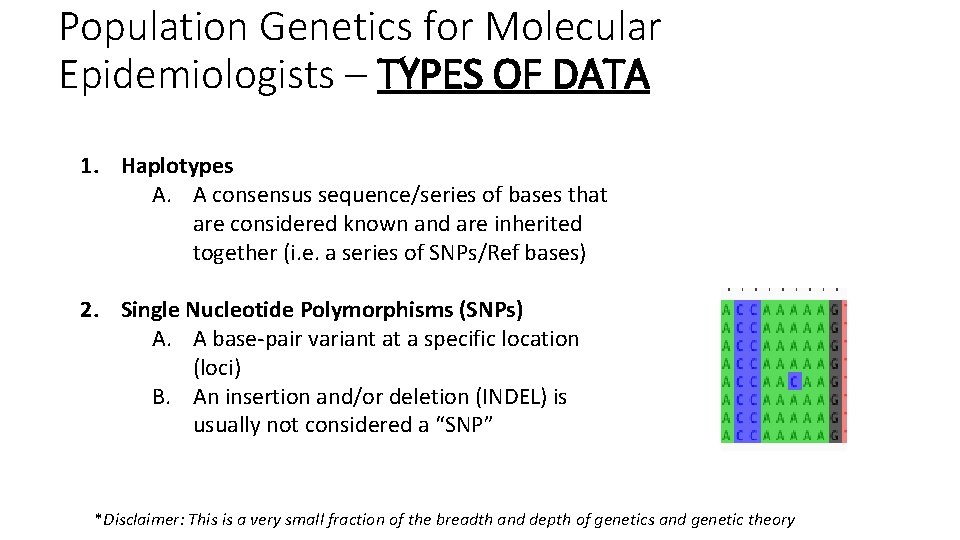
Population Genetics for Molecular Epidemiologists – TYPES OF DATA 1. Haplotypes A. A consensus sequence/series of bases that are considered known and are inherited together (i. e. a series of SNPs/Ref bases) 2. Single Nucleotide Polymorphisms (SNPs) A. A base-pair variant at a specific location (loci) B. An insertion and/or deletion (INDEL) is usually not considered a “SNP” *Disclaimer: This is a very small fraction of the breadth and depth of genetics and genetic theory
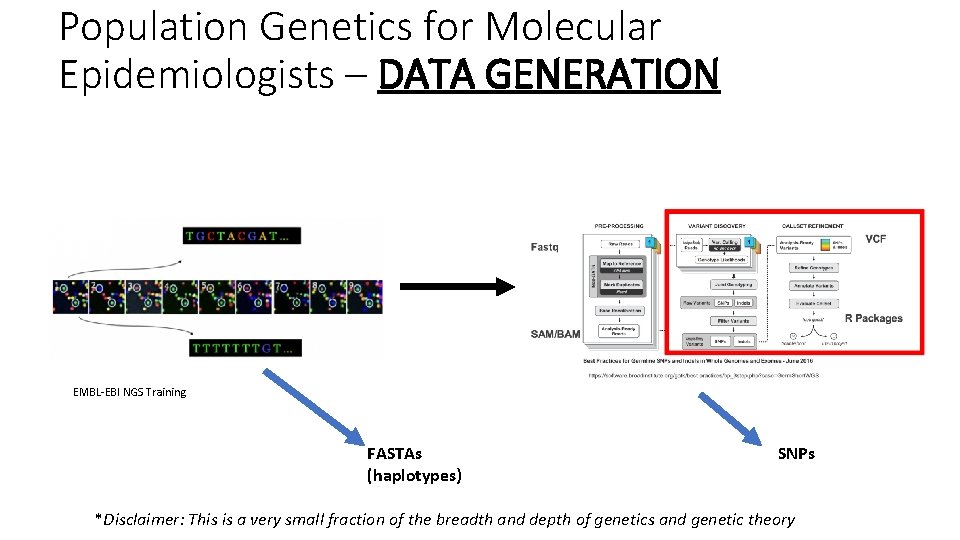
Population Genetics for Molecular Epidemiologists – DATA GENERATION EMBL-EBI NGS Training FASTAs (haplotypes) SNPs *Disclaimer: This is a very small fraction of the breadth and depth of genetics and genetic theory
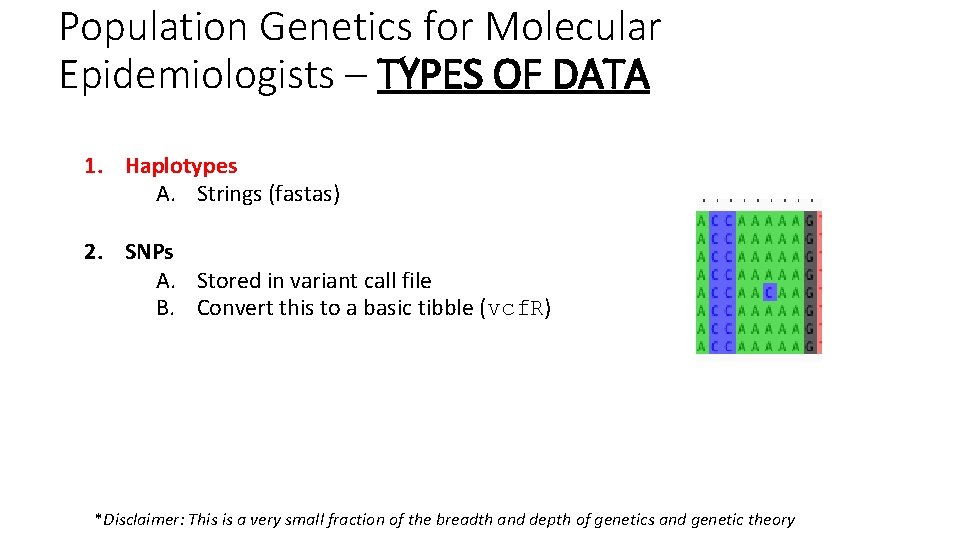
Population Genetics for Molecular Epidemiologists – TYPES OF DATA 1. Haplotypes A. Strings (fastas) 2. SNPs A. Stored in variant call file B. Convert this to a basic tibble (vcf. R) *Disclaimer: This is a very small fraction of the breadth and depth of genetics and genetic theory

Haplotypes • Strings » ATCG APE package is popular for phylogenetic analysis and manipulations • Very efficient storage (DNA as bytes)
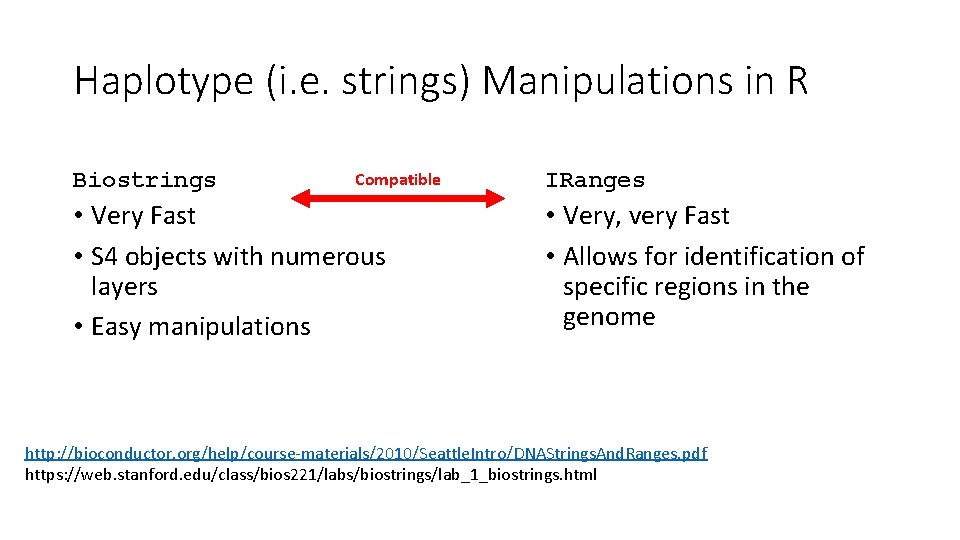
Haplotype (i. e. strings) Manipulations in R Biostrings Compatible • Very Fast • S 4 objects with numerous layers • Easy manipulations IRanges • Very, very Fast • Allows for identification of specific regions in the genome http: //bioconductor. org/help/course-materials/2010/Seattle. Intro/DNAStrings. And. Ranges. pdf https: //web. stanford. edu/class/bios 221/labs/biostrings/lab_1_biostrings. html
Population Genetics for Molecular Epidemiologists – TYPES OF DATA 1. Haplotypes A. Strings (fastas) 2. SNPs A. Stored in variant call file B. Convert this to a basic tibble (vcf. R) *Disclaimer: This is a very small fraction of the breadth and depth of genetics and genetic theory

Variant Call Files Show on board as well
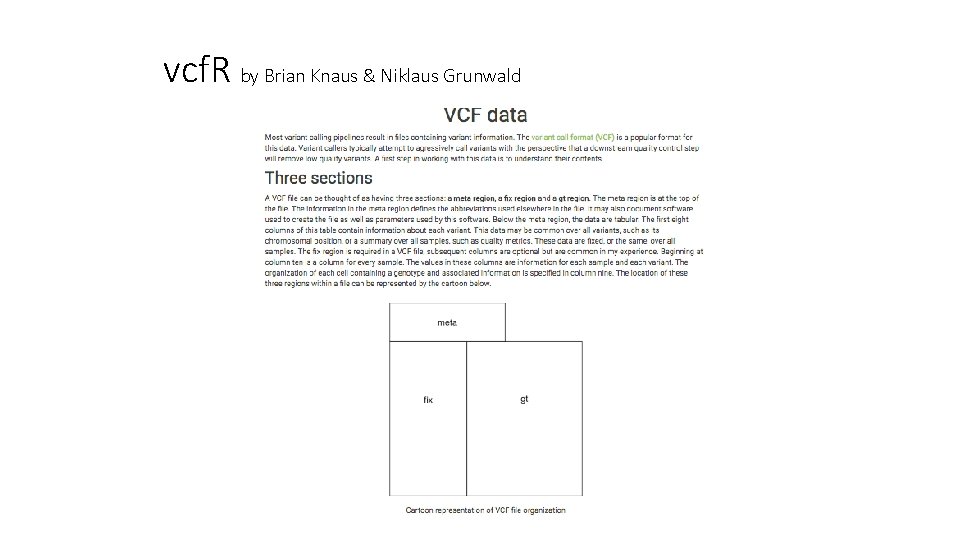
vcf. R by Brian Knaus & Niklaus Grunwald
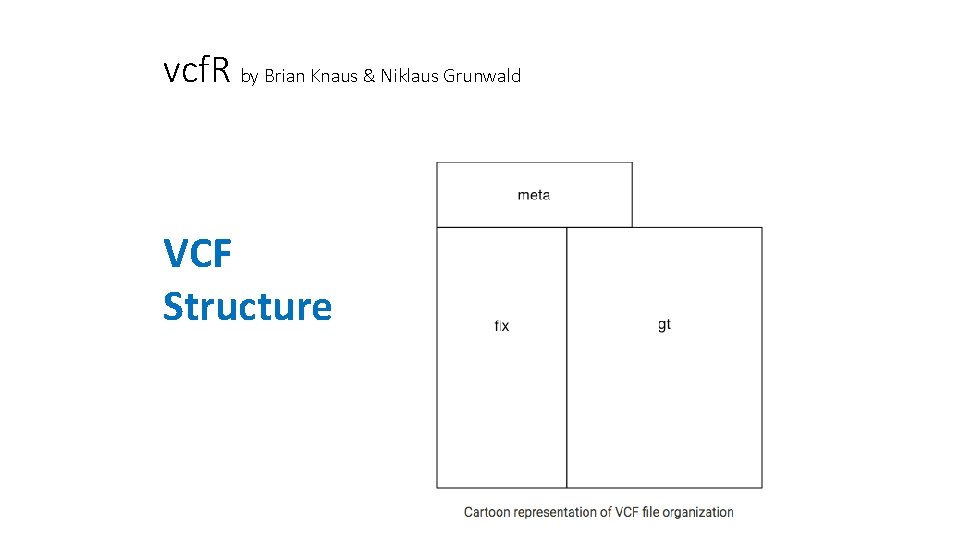
vcf. R by Brian Knaus & Niklaus Grunwald VCF Structure
poppr by Niklaus Grunwald 1. Open R (Rstudio) 2. install. packages(“poppr”) 3. Online textbook & tutorials by Grunwald Lab

poppr by Niklaus Grunwald 1. Do all parts of this Population genetics and genomics in R primer/textbook (because it is great information/work)! A. But especially high-yield is Part III
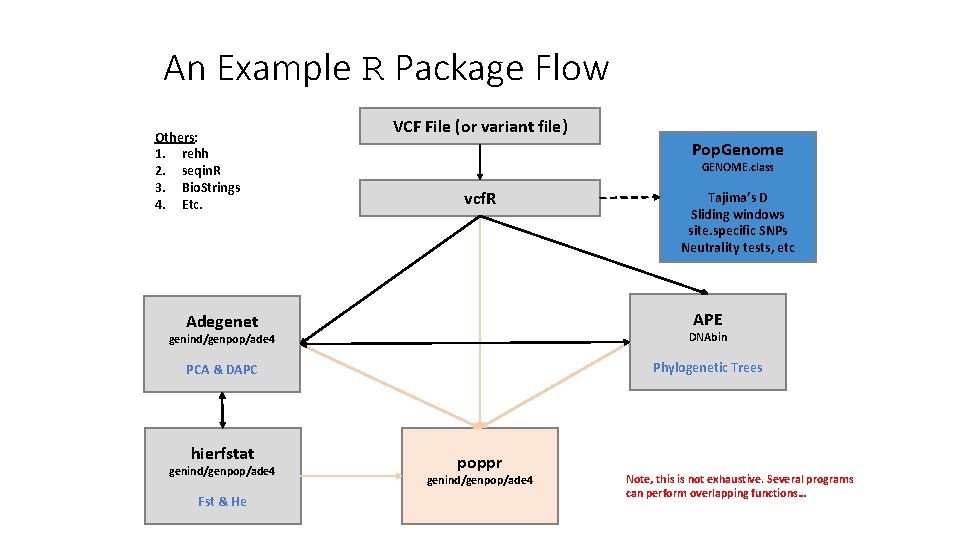
An Example R Package Flow Others: 1. rehh 2. seqin. R 3. Bio. Strings 4. Etc. VCF File (or variant file) Pop. Genome GENOME. class vcf. R Tajima’s D Sliding windows site. specific SNPs Neutrality tests, etc APE Adegenet genind/genpop/ade 4 DNAbin PCA & DAPC Phylogenetic Trees hierfstat genind/genpop/ade 4 Fst & He poppr genind/genpop/ade 4 Note, this is not exhaustive. Several programs can perform overlapping functions…

Plan • Rmarkdown example

R Pop Gen Resources 1. poppr Textbook/Primer 2. NESCENT Hackathon A. B. https: //github. com/NESCent/r-popgen-hackathon popgen. nescent. org/ 3. Package Specific VIGNETTES, Tutorials, & Manuals 4. Molecular Ecology Resources: Review of Pop. Gen in R (http: //onlinelibrary. wiley. com/doi/10. 1111/men. 2017. issue-1/issuetoc) 5. Computational Genome Analysis (textbook) by Richard Deonier, Simon Tavare, Michael Waterman
- Slides: 43