Stochastic Roadmap Simulation An Efficient Representation and Algorithm




- Slides: 14

Stochastic Roadmap Simulation: An Efficient Representation and Algorithm for Analyzing Molecular Motion Mehmet Serkan Apaydin, Douglas L. Brutlag, Carlos Guestrin, David Hsu, Jean-Claude Latombe Presented by: Alan Chen

Outline Introduction n Stochastic Roadmap Simulation (SRS) n First-step Analysis and Roadmap Query n SRS vs. Monte Carlo n Transmission Coefficients n Results n Discussions n

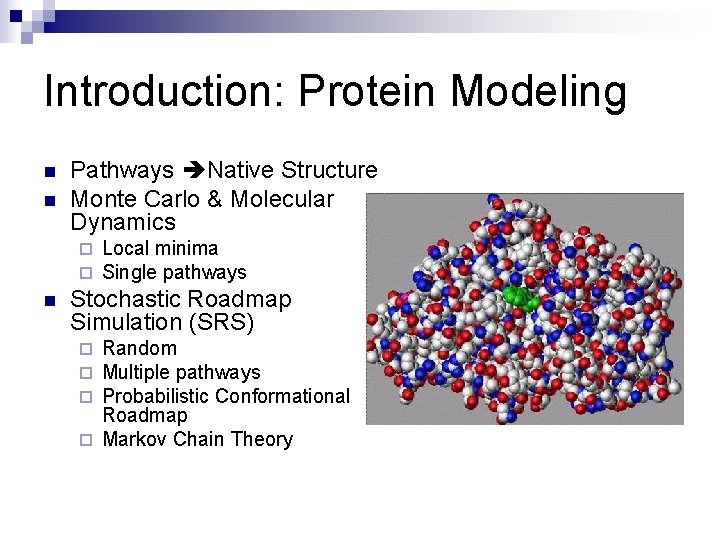
Introduction: Protein Modeling n n Pathways Native Structure Monte Carlo & Molecular Dynamics ¨ ¨ n Local minima Single pathways Stochastic Roadmap Simulation (SRS) Random Multiple pathways Probabilistic Conformational Roadmap ¨ Markov Chain Theory ¨ ¨ ¨

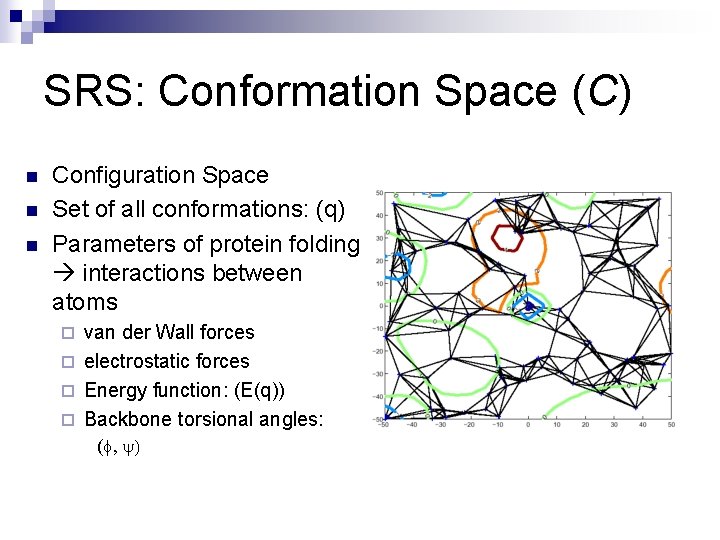
SRS: Conformation Space (C) n n n Configuration Space Set of all conformations: (q) Parameters of protein folding interactions between atoms van der Wall forces ¨ electrostatic forces ¨ Energy function: (E(q)) ¨ Backbone torsional angles: ¨ (f, y)

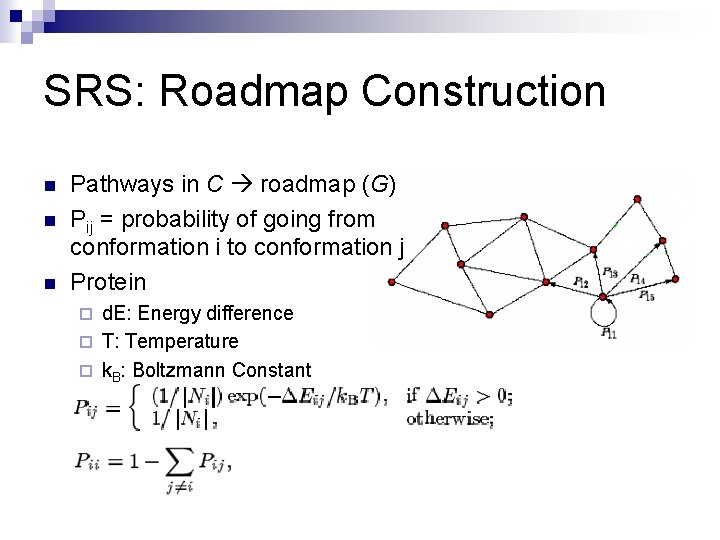
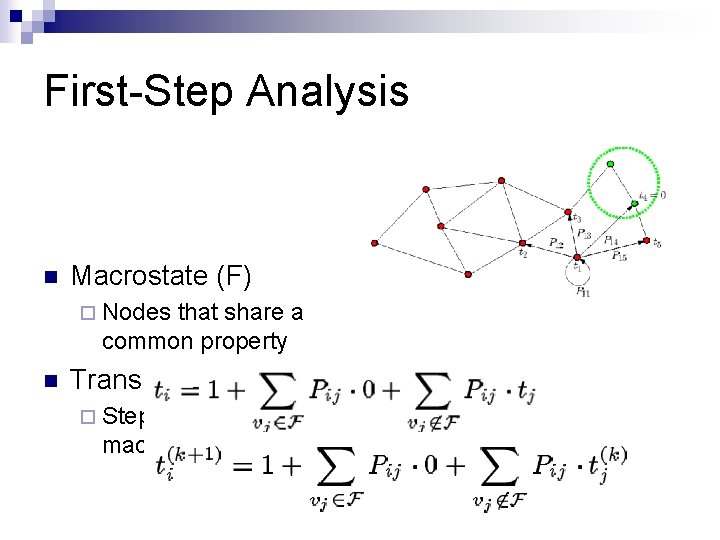
SRS: Roadmap Construction n Pathways in C roadmap (G) Pij = probability of going from conformation i to conformation j Protein d. E: Energy difference ¨ T: Temperature ¨ k. B: Boltzmann Constant ¨

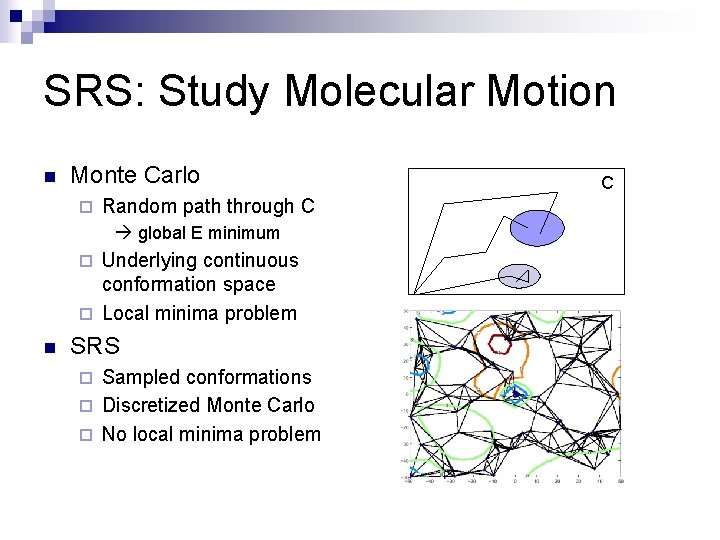
SRS: Study Molecular Motion n Monte Carlo ¨ Random path through C global E minimum Underlying continuous conformation space ¨ Local minima problem ¨ n SRS Sampled conformations ¨ Discretized Monte Carlo ¨ No local minima problem ¨ C

First-Step Analysis n Macrostate (F) ¨ Nodes that share a common property n Transitions (t) ¨ Steps from a node to a macrostate

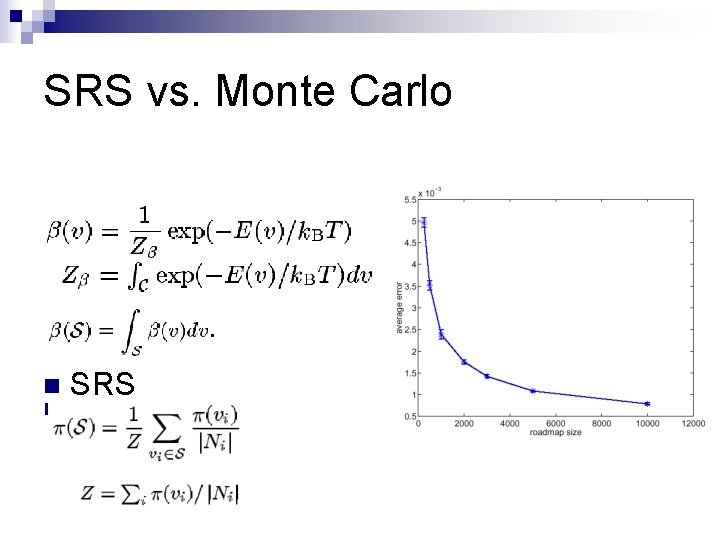
SRS vs. Monte Carlo Associated limiting distribution p n Stationary distribution n = Spj. Pji ¨ pi > 0 ¨ pi ¨S p i = 1 p 2 p 3 p 1

SRS vs. Monte Carlo n n SRS Monte Carlo

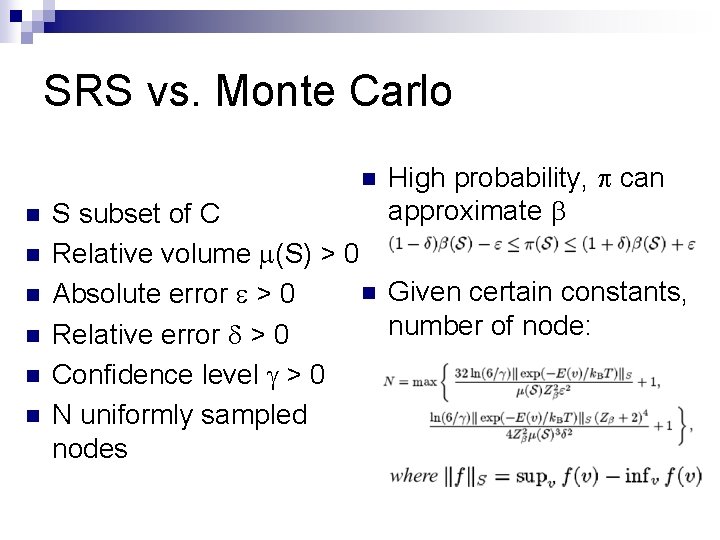
SRS vs. Monte Carlo n n n n High probability, p can approximate b S subset of C Relative volume m(S) > 0 n Given certain constants, Absolute error e > 0 number of node: Relative error d > 0 Confidence level g > 0 N uniformly sampled nodes

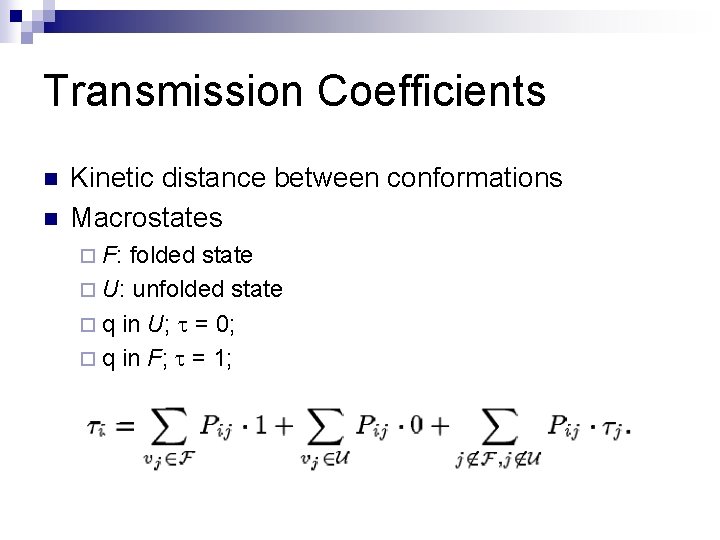
Transmission Coefficients n n Kinetic distance between conformations Macrostates ¨ F: folded state ¨ U: unfolded state ¨ q in U; t = 0; ¨ q in F; t = 1;

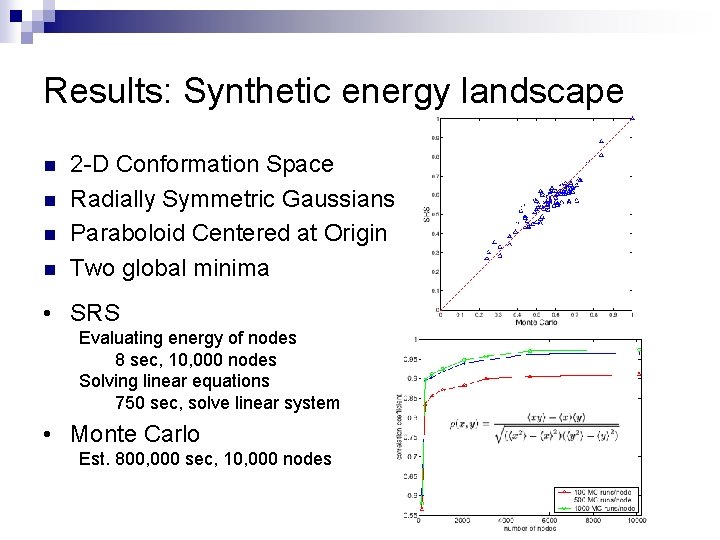
Results: Synthetic energy landscape n n 2 -D Conformation Space Radially Symmetric Gaussians Paraboloid Centered at Origin Two global minima • SRS Evaluating energy of nodes 8 sec, 10, 000 nodes Solving linear equations 750 sec, solve linear system • Monte Carlo Est. 800, 000 sec, 10, 000 nodes

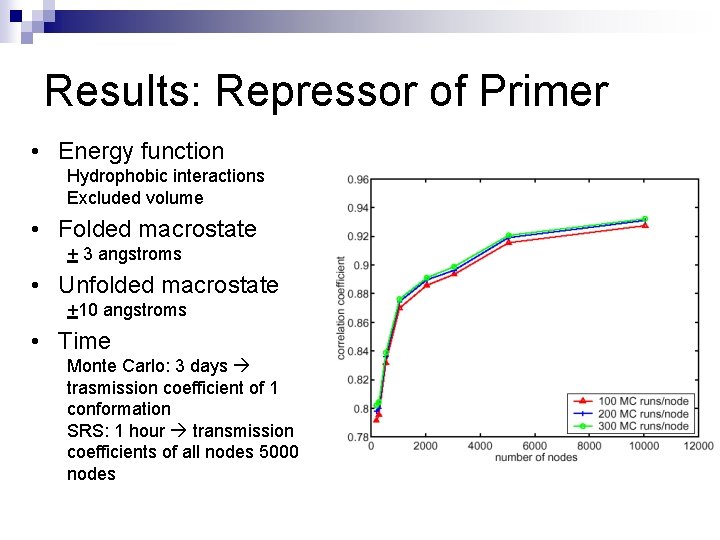
Results: Repressor of Primer • Energy function Hydrophobic interactions Excluded volume • Folded macrostate + 3 angstroms • Unfolded macrostate +10 angstroms • Time Monte Carlo: 3 days trasmission coefficient of 1 conformation SRS: 1 hour transmission coefficients of all nodes 5000 nodes

Discussions n SRS vs Monte Carlo ¨ ¨ ¨ n multiple paths vs. single path In the limit, SRS converges to Monte Carlo One hour vs. three days Improvements ¨ Better roadmaps n n ¨ n Reduce the dimension of C Better sampling strategy Faster linear system solver Uses ¨ ¨ Order of protein folding Overcoming energy barriers (catalytic sites)