Statistical Power Calculations Manuel AR Ferreira Massachusetts General
















- Slides: 39

Statistical Power Calculations Manuel AR Ferreira Massachusetts General Hospital Harvard Medical School Boston Boulder, 2007

Outline 1. Aim 2. Statistical power 3. Estimate the power of linkage / association analysis Analytically Empirically 4. Improve the power of linkage analysis

1. Aim

1. Know what type-I error and power are 2. Know that you can/should estimate the power of your linkage/association analyses (analytically or empirically) 3. Know that there a number of tools that you can use to estimate power 4. Be aware that there are MANY factors that increase type-I error and decrease power

2. Statistical power

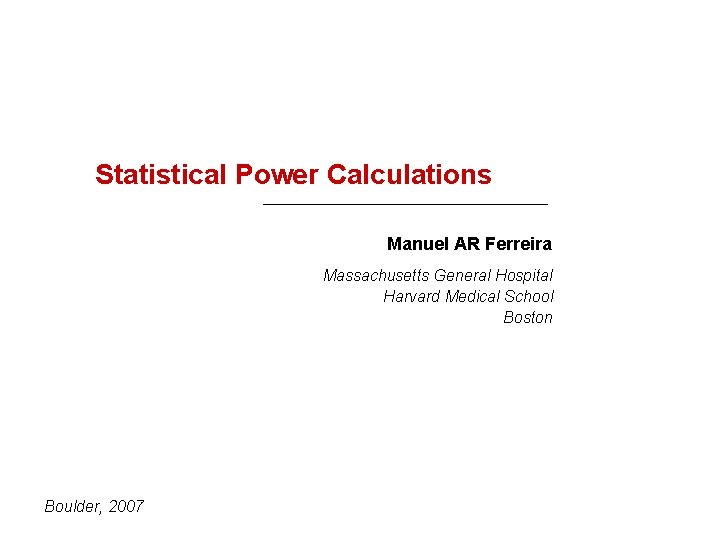
H 0: Person A is not guilty H 1: Person A is guilty – send him to jail In reality… We decide… H 0 is true H 1 is true 1 -α H 1 is true β Type-2 error α 1 -β Type-1 error Power: probability of declaring that something is true when in reality it is true.

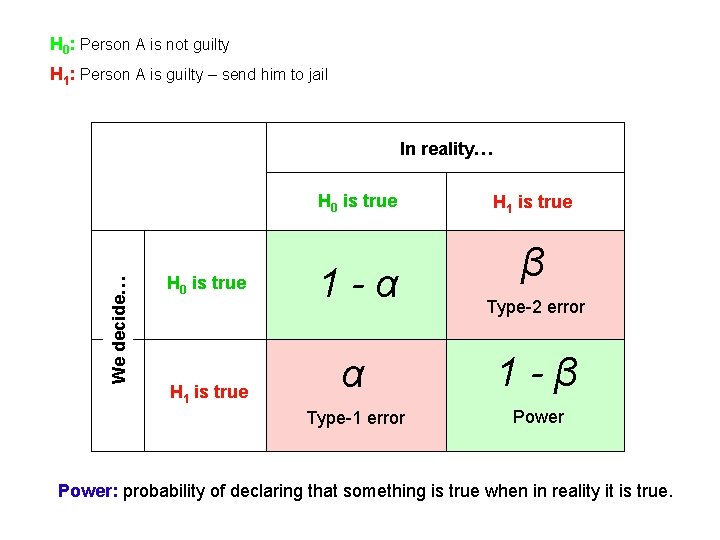
H 0: There is NO linkage between a marker and a trait H 1: There is linkage between a marker and a trait Linkage test statistic has different distributions under H 0 and H 1 xx xx xxx x x xxx xxx xx x x xx x x xx x

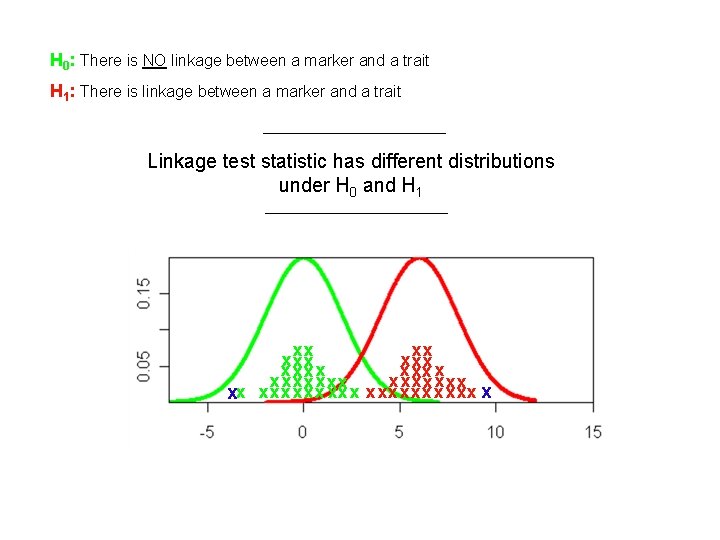
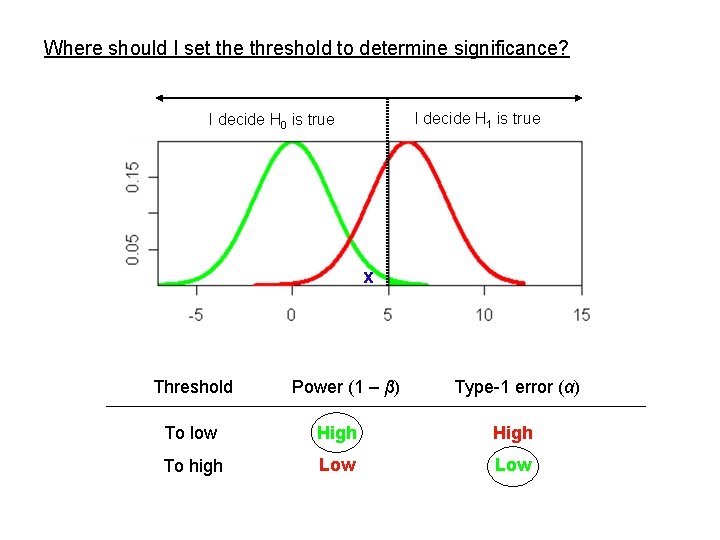
Where should I set the threshold to determine significance? I decide H 0 is true I decide H 1 is true (Linkage) x Threshold To low Power (1 – β) High Type-1 error (α) High

Where should I set the threshold to determine significance? I decide H 1 is true I decide H 0 is true x Threshold Power (1 – β) Type-1 error (α) To low High To high Low

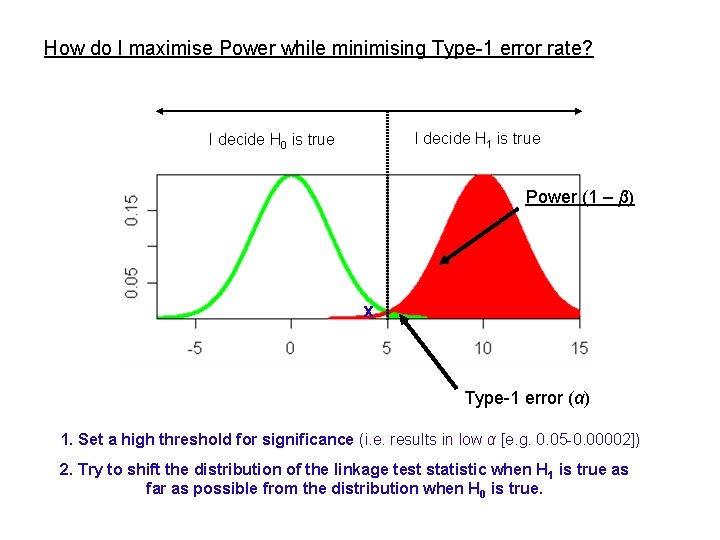
How do I maximise Power while minimising Type-1 error rate? I decide H 1 is true I decide H 0 is true Power (1 – β) x Type-1 error (α) 1. Set a high threshold for significance (i. e. results in low α [e. g. 0. 05 -0. 00002]) 2. Try to shift the distribution of the linkage test statistic when H 1 is true as far as possible from the distribution when H 0 is true.

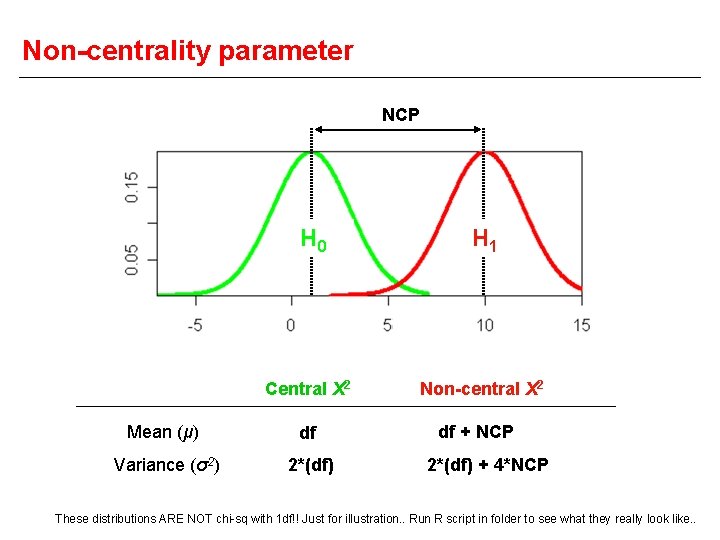
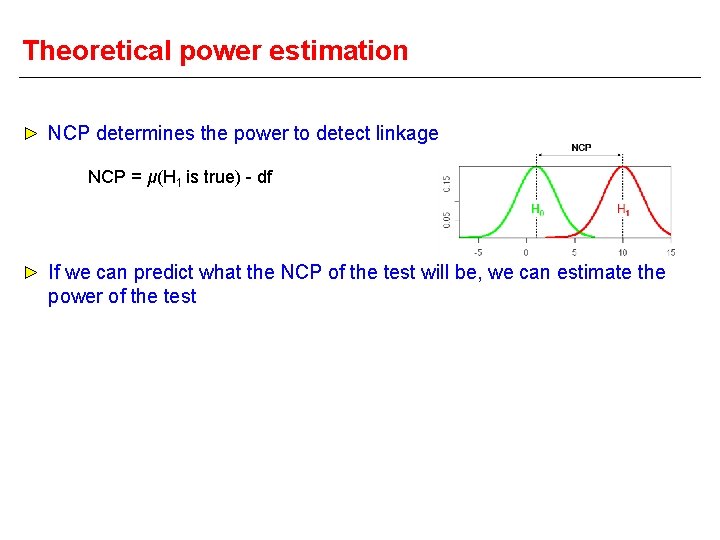
Non-centrality parameter NCP H 0 Central Χ 2 Mean (μ) df Variance (σ2) 2*(df) H 1 Non-central Χ 2 df + NCP 2*(df) + 4*NCP These distributions ARE NOT chi-sq with 1 df!! Just for illustration. . Run R script in folder to see what they really look like. .

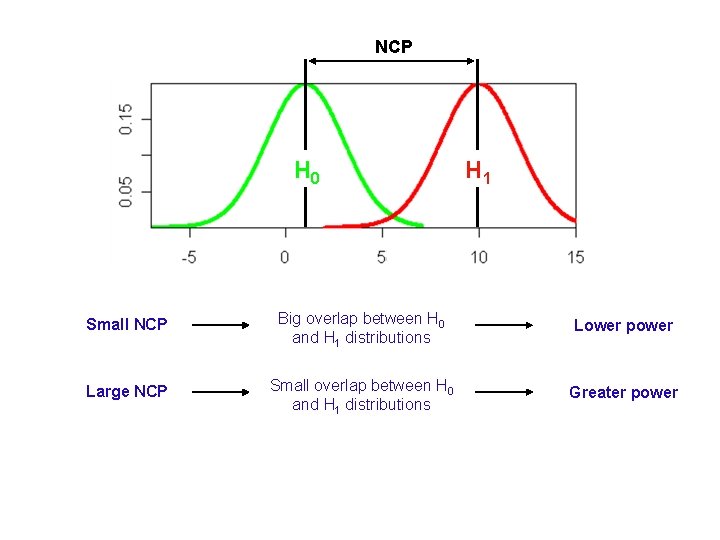
NCP H 0 H 1 Small NCP Big overlap between H 0 and H 1 distributions Lower power Large NCP Small overlap between H 0 and H 1 distributions Greater power

Short practical on GPC Genetic Power Calculator is an online resource for carrying out basic power calculations. http: //pngu. mgh. harvard. edu/~purcell/gpc/ For our 1 st example we will use the probability function calculator to play with power

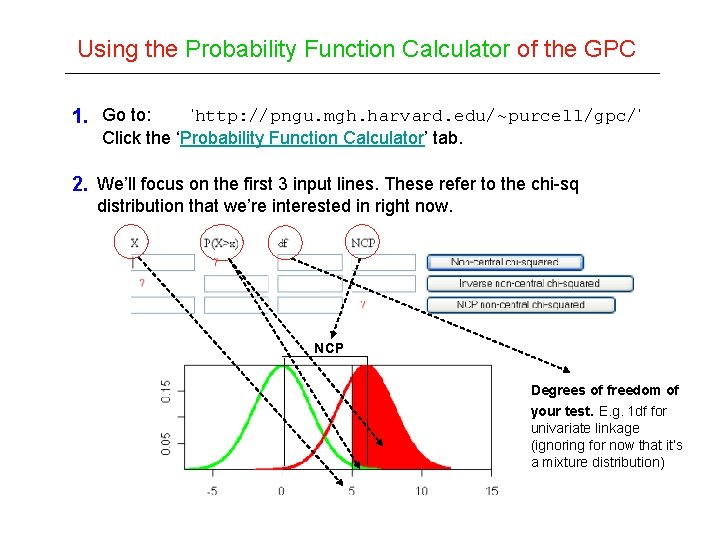
Using the Probability Function Calculator of the GPC 1. Go to: ‘http: //pngu. mgh. harvard. edu/~purcell/gpc/’ Click the ‘Probability Function Calculator’ tab. 2. We’ll focus on the first 3 input lines. These refer to the chi-sq distribution that we’re interested in right now. NCP Degrees of freedom of your test. E. g. 1 df for univariate linkage (ignoring for now that it’s a mixture distribution)

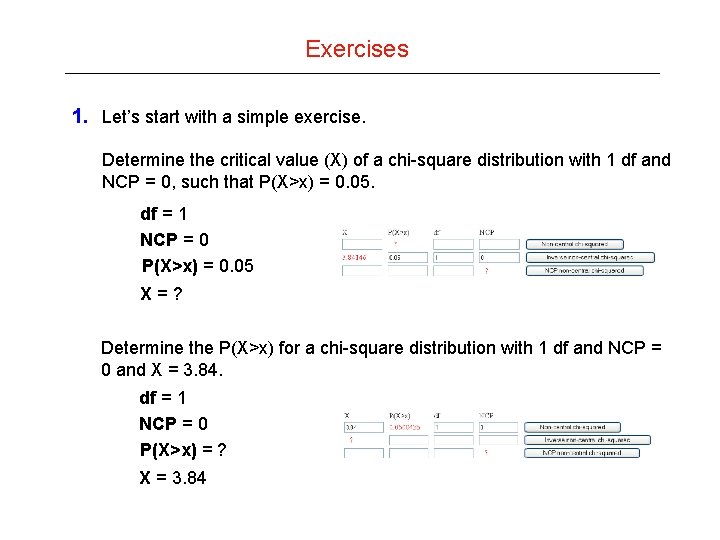
Exercises 1. Let’s start with a simple exercise. Determine the critical value (X) of a chi-square distribution with 1 df and NCP = 0, such that P(X>x) = 0. 05. df = 1 NCP = 0 P(X>x) = 0. 05 X=? Determine the P(X>x) for a chi-square distribution with 1 df and NCP = 0 and X = 3. 84. df = 1 NCP = 0 P(X>x) = ? X = 3. 84

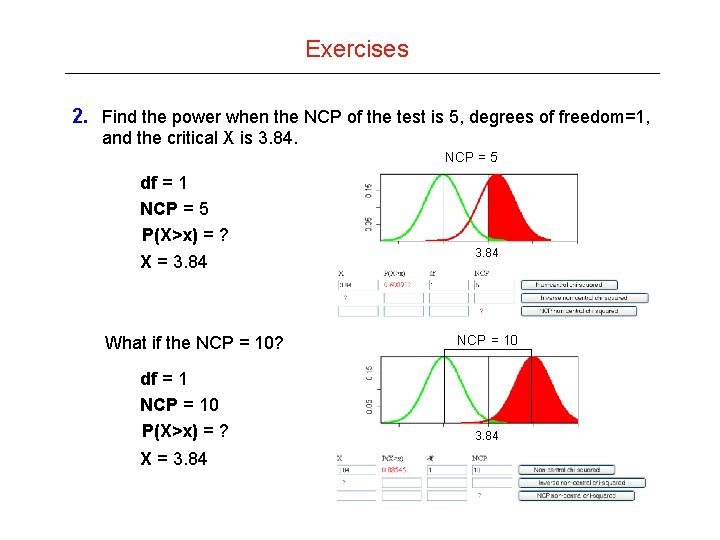
Exercises 2. Find the power when the NCP of the test is 5, degrees of freedom=1, and the critical X is 3. 84. NCP = 5 df = 1 NCP = 5 P(X>x) = ? X = 3. 84 What if the NCP = 10? df = 1 NCP = 10 P(X>x) = ? X = 3. 84 NCP = 10 3. 84

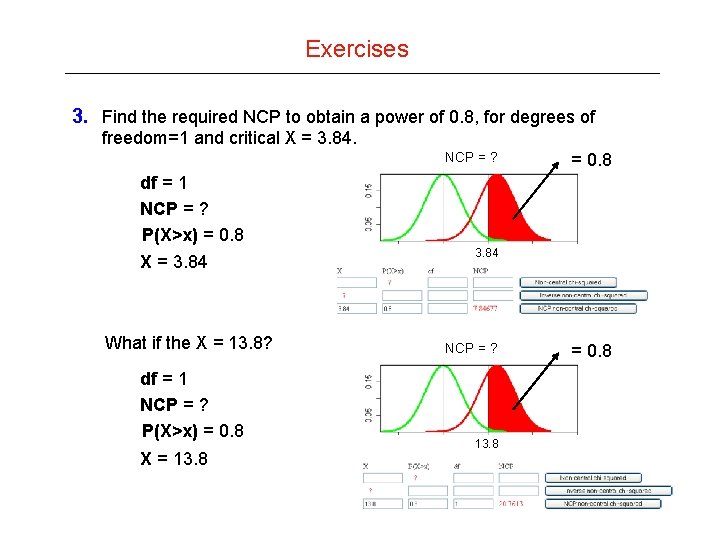
Exercises 3. Find the required NCP to obtain a power of 0. 8, for degrees of freedom=1 and critical X = 3. 84. NCP = ? = 0. 8 df = 1 NCP = ? P(X>x) = 0. 8 X = 3. 84 What if the X = 13. 8? df = 1 NCP = ? P(X>x) = 0. 8 X = 13. 84 NCP = ? 13. 8 = 0. 8

2. Estimate power for linkage and association

Why is it important to estimate power? To determine whether the study you’re designing/analysing can in fact localise the QTL you’re looking for. Study design and interpretation of results. You’ll need to do it for most grant applications. When and how should I estimate power? When? How? Study design stage Theoretically, empirically Analysis stage Empirically

Theoretical power estimation NCP determines the power to detect linkage NCP = μ(H 1 is true) - df If we can predict what the NCP of the test will be, we can estimate the power of the test

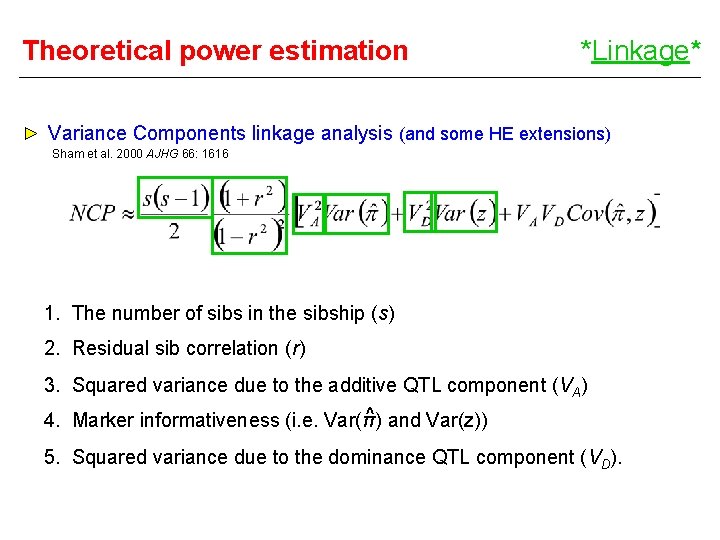
Theoretical power estimation *Linkage* Variance Components linkage analysis (and some HE extensions) Sham et al. 2000 AJHG 66: 1616 1. The number of sibs in the sibship (s) 2. Residual sib correlation (r) 3. Squared variance due to the additive QTL component (VA) ^ and Var(z)) 4. Marker informativeness (i. e. Var(π) 5. Squared variance due to the dominance QTL component (VD).

Another short practical on GPC The idea is to see how genetic parameters and the study design influence the NCP – and so the power – of linkage analysis

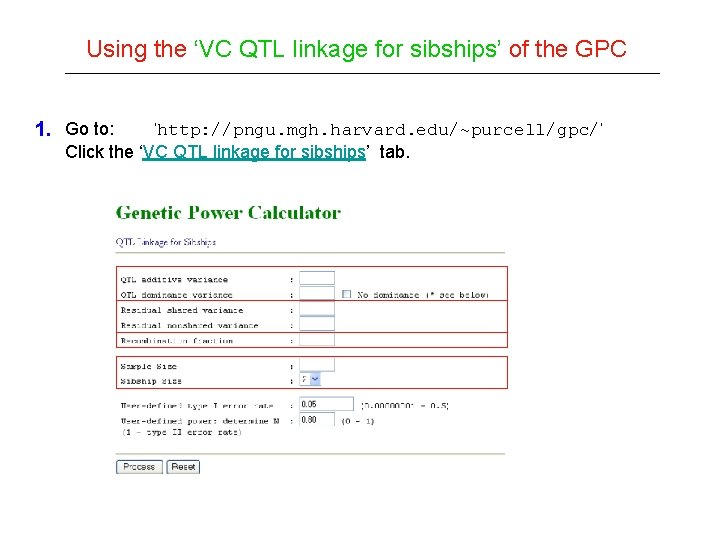
Using the ‘VC QTL linkage for sibships’ of the GPC 1. Go to: ‘http: //pngu. mgh. harvard. edu/~purcell/gpc/’ Click the ‘VC QTL linkage for sibships’ tab.

Exercises 1. Let’s estimate the power of linkage for the following parameters: QTL additive variance: 0. 2 QTL dominance variance: 0 Residual shared variance: 0. 4 Residual nonshared variance: Recombination fraction: Sample Size: 200 Sibship Size: 2 0. 4 0 User-defined type I error rate: 0. 05 User-defined power: determine N : 0. 8 Power = 0. 36 (alpha = 0. 05) Sample size for 80% power = 681 families

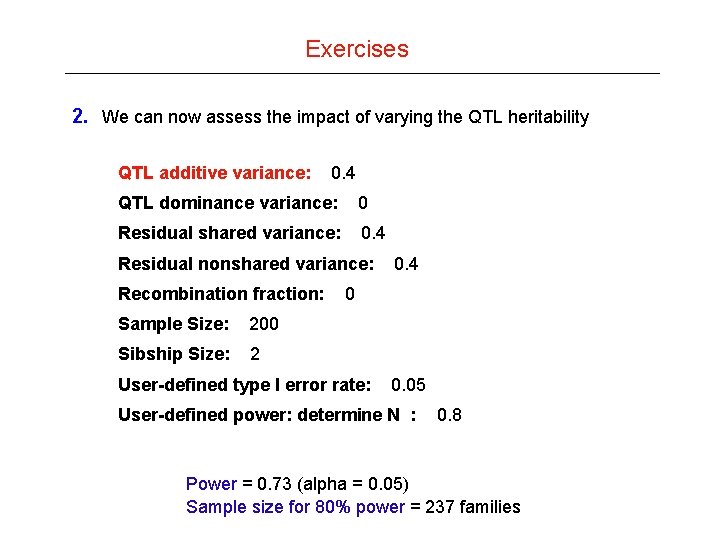
Exercises 2. We can now assess the impact of varying the QTL heritability QTL additive variance: 0. 4 QTL dominance variance: 0 Residual shared variance: 0. 4 Residual nonshared variance: Recombination fraction: Sample Size: 200 Sibship Size: 2 0. 4 0 User-defined type I error rate: 0. 05 User-defined power: determine N : 0. 8 Power = 0. 73 (alpha = 0. 05) Sample size for 80% power = 237 families

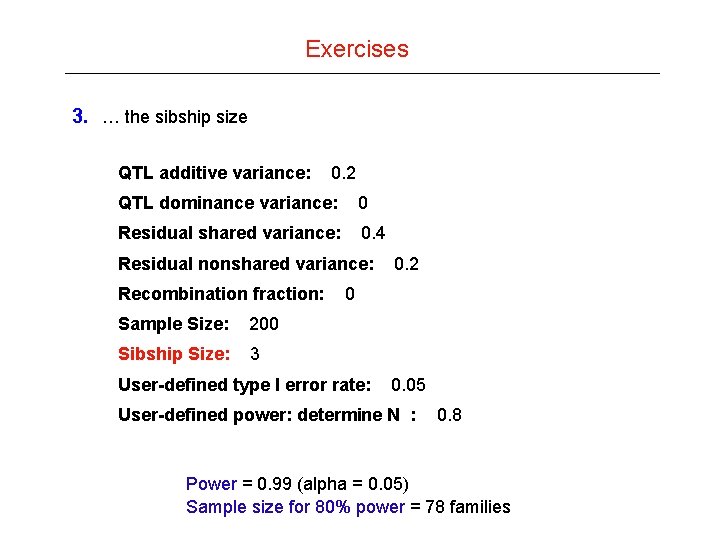
Exercises 3. … the sibship size QTL additive variance: 0. 2 QTL dominance variance: 0 Residual shared variance: 0. 4 Residual nonshared variance: Recombination fraction: Sample Size: 200 Sibship Size: 3 0. 2 0 User-defined type I error rate: 0. 05 User-defined power: determine N : 0. 8 Power = 0. 99 (alpha = 0. 05) Sample size for 80% power = 78 families

Theoretical power estimation *Association: case-control* Ca. TS performs power calculations for large genetic association studies, including two stage studies. http: //www. sph. umich. edu/csg/abecasis/Ca. TS/index. html

Theoretical power estimation *Association: TDT* TDT Power calculator, while accounting for the effects of untested loci and shared environmental factors that also contribute to disease risk http: //pngu. mgh. harvard. edu/~mferreira/power_tdt/calculator. html

Theoretical power estimation Advantages: Fast, GPC, Ca. TS Disadvantages: Approximation, may not fit well individual study designs, particularly if one needs to consider more complex pedigrees, missing data, ascertainment strategies, different tests, etc…

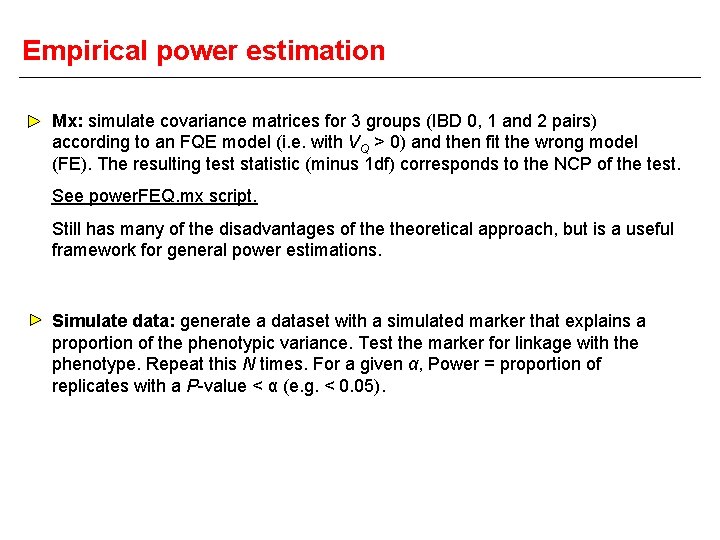
Empirical power estimation Mx: simulate covariance matrices for 3 groups (IBD 0, 1 and 2 pairs) according to an FQE model (i. e. with VQ > 0) and then fit the wrong model (FE). The resulting test statistic (minus 1 df) corresponds to the NCP of the test. See power. FEQ. mx script. Still has many of the disadvantages of theoretical approach, but is a useful framework for general power estimations. Simulate data: generate a dataset with a simulated marker that explains a proportion of the phenotypic variance. Test the marker for linkage with the phenotype. Repeat this N times. For a given α, Power = proportion of replicates with a P-value < α (e. g. < 0. 05).

Empirical power estimation *Linkage / Association* Example with LINX http: //pngu. mgh. harvard. edu/~mferreira/

3. How to improve power

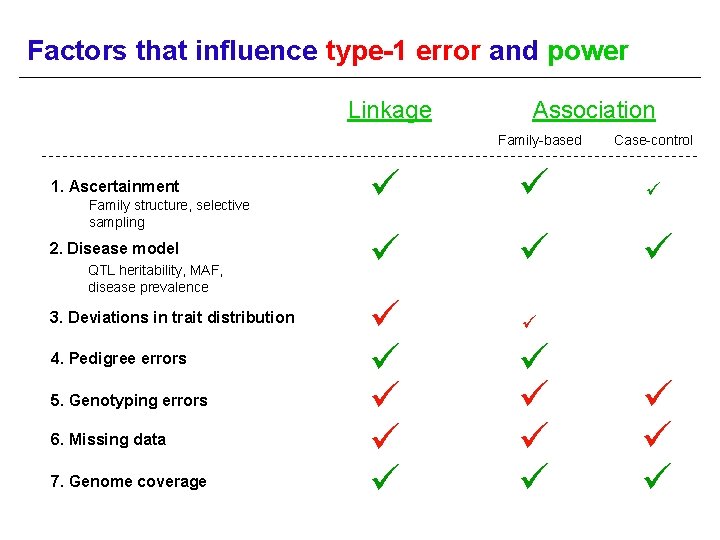
Factors that influence type-1 error and power Linkage Association Family-based 1. Ascertainment Family structure, selective sampling 2. Disease model QTL heritability, MAF, disease prevalence 3. Deviations in trait distribution 4. Pedigree errors 5. Genotyping errors 6. Missing data 7. Genome coverage Case-control

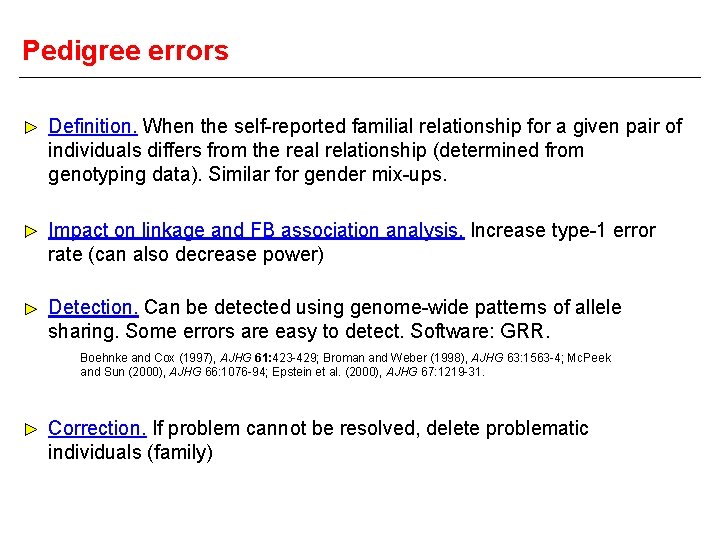
Pedigree errors Definition. When the self-reported familial relationship for a given pair of individuals differs from the real relationship (determined from genotyping data). Similar for gender mix-ups. Impact on linkage and FB association analysis. Increase type-1 error rate (can also decrease power) Detection. Can be detected using genome-wide patterns of allele sharing. Some errors are easy to detect. Software: GRR. Boehnke and Cox (1997), AJHG 61: 423 -429; Broman and Weber (1998), AJHG 63: 1563 -4; Mc. Peek and Sun (2000), AJHG 66: 1076 -94; Epstein et al. (2000), AJHG 67: 1219 -31. Correction. If problem cannot be resolved, delete problematic individuals (family)

Pedigree errors *Impact on linkage* • CSGA (1997) A genome-wide search for asthma susceptibility loci in ethnically diverse populations. Nat Genet 15: 389 -92 • ~15 families with wrong relationships • No significant evidence for linkage • Error checking is essential!


Pedigree errors GRR *Detection/Correction* http: //www. sph. umich. edu/csg/abecasis

Practical Aim: Identify pedigree errors with GRR 1. Go to: ‘EgmondserversharePrograms’ Copy entire ‘GRR’ folder into your desktop. 2. Go into the ‘GRR’ folder in your desktop, and run the GRR. exe file. 3. Press the ‘Load’ button, and navigate into the same ‘GRR’ folder on the desktop. Select the file ‘sample. ped’ and press ‘Open’. Note that all sibpairs in ‘sample. ped’ were reported to be fullsibs or half-sibs. I’ll identify one error. Can you identify the other two?

Summmary 1. Statistical power 2. Estimate the power of linkage analysis 3. Improve the power of linkage analysis