Statistical Methods for Data Analysis a Roo Stats



![Replace nsig by σ×ε×L // signal yield // p. Ws->factory( "nsig[0, 0, 100]" ); Replace nsig by σ×ε×L // signal yield // p. Ws->factory( "nsig[0, 0, 100]" );](https://slidetodoc.com/presentation_image_h2/c0af792ad71784add92df5de1fc9ad61/image-6.jpg)



- Slides: 18

Statistical Methods for Data Analysis a Roo. Stats example Luca Lista INFN Napoli

Roo. Stats toolkit • Concepts: – PDF modeling: done via Roo. Fit package – Workspace: an area where the PDF and data model can be defined, and saved to disk for later use – Interval Calculator: abstract class for computation of confidence intervals: • Bayesian (plain, Markov Chain), central Neyman, Feldman-Cousins, … – Hypothesis test calculator: abstract class to compute p-values, significance, CLs, … Luca Lista Statistical Methods for Data Analysis 2

Step-by-step example • Example presented during the last CMS Data Analysis School (FNAL-Pisa), by Gena Kukartsev • Create a ROOT macro, say counting. C, with a void function, say Make. Workspace() – #include directives and other details skipped for sake of simplicity; complete code available on request • Create workspace, save to disk void Make. Workspace( void ){ // create workspace Roo. Workspace * p. Ws = new Roo. Workspace("my. WS"); // save workspace to file p. Ws->Save. As("workspace. root”); return; } Luca Lista Statistical Methods for Data Analysis 3

Define parameters and PDF model • Create workspace // create workspace Roo. Workspace * p. Ws = new Roo. Workspace("my. WS"); // observable: number of events p. Ws->factory( "n[0]" ); // signal yield p. Ws->factory( "nsig[0, 0, 100]" ); // NOTE: three parameters are "current value", "low bound", "upper bound” // background yield p. Ws->factory( "nbkg[10, 0, 100]" ); // full event yield p. Ws->factory( "sum: : yield(nsig, nbkg)" ); // NOTE: lower-case "sum" create a function. Upper-case "SUM" would create a PDF // Core model: Poisson probability with mean signal+bkg p. Ws->factory( "Poisson: : model_core(n, yield) " ); // NOTE: "model_core" is a name of the PDF object // print out the workspace contents p. Ws->Print(); // save workspace to file p. Ws->Save. As("workspace. root”); Luca Lista Statistical Methods for Data Analysis 4

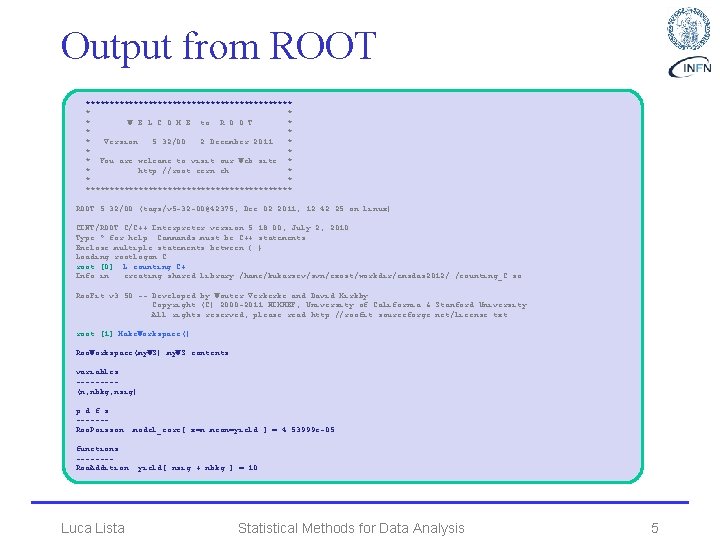
Output from ROOT ********************** * W E L C O M E to R O O T * * Version 5. 32/00 2 December 2011 * * You are welcome to visit our Web site * * http: //root. cern. ch * ********************** ROOT 5. 32/00 (tags/v 5 -32 -00@42375, Dec 02 2011, 12: 42: 25 on linux) CINT/ROOT C/C++ Interpreter version 5. 18. 00, July 2, 2010 Type ? for help. Commands must be C++ statements. Enclose multiple statements between { }. Loading rootlogon. C. . . root [0]. L counting. C+ Info in : creating shared library /home/kukarzev/svn/exost/workdir/cmsdas 2012/. /counting_C. so Roo. Fit v 3. 50 -- Developed by Wouter Verkerke and David Kirkby Copyright (C) 2000 -2011 NIKHEF, University of California & Stanford University All rights reserved, please read http: //roofit. sourceforge. net/license. txt root [1] Make. Workspace() Roo. Workspace(my. WS) my. WS contents variables ----(n, nbkg, nsig) p. d. f. s ------Roo. Poisson: : model_core[ x=n mean=yield ] = 4. 53999 e-05 functions -------Roo. Addition: : yield[ nsig + nbkg ] = 10 Luca Lista Statistical Methods for Data Analysis 5
![Replace nsig by σεL signal yield p Wsfactory nsig0 0 100 Replace nsig by σ×ε×L // signal yield // p. Ws->factory( "nsig[0, 0, 100]" );](https://slidetodoc.com/presentation_image_h2/c0af792ad71784add92df5de1fc9ad61/image-6.jpg)
Replace nsig by σ×ε×L // signal yield // p. Ws->factory( "nsig[0, 0, 100]" ); // integrated luminosity p. Ws->factory( "lumi[0]" ); // cross section - parameter of interest p. Ws->factory( "xsec[0, 0, 0. 1]" ); // selection efficiency * acceptance p. Ws->factory( "efficiency[0]" ); // signal yield p. Ws->factory( "prod: : nsig(lumi, xsec, efficiency)" ); Define prior PDF (uniform) // define Bayesian prior PDF for POI p. Ws->factory( "Uniform: : prior(xsec)" ); Luca Lista Statistical Methods for Data Analysis 6

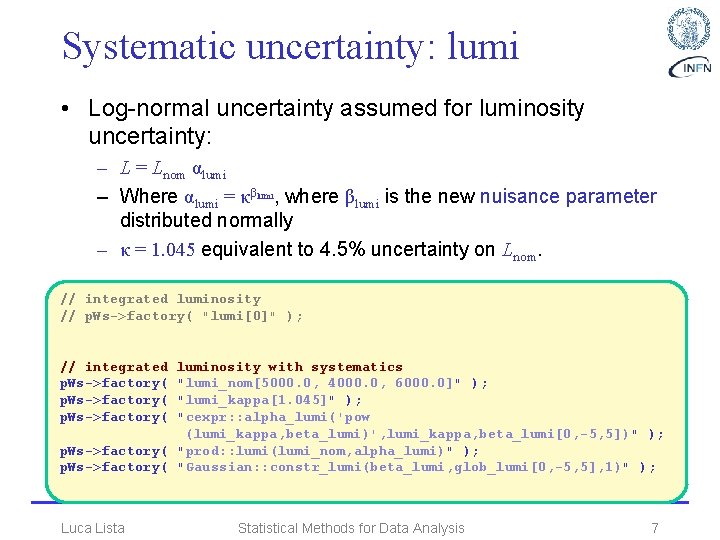
Systematic uncertainty: lumi • Log-normal uncertainty assumed for luminosity uncertainty: – L = Lnom αlumi – Where αlumi = κβlumi, where βlumi is the new nuisance parameter distributed normally – κ = 1. 045 equivalent to 4. 5% uncertainty on Lnom. // integrated luminosity // p. Ws->factory( "lumi[0]" ); // integrated p. Ws->factory( luminosity with systematics "lumi_nom[5000. 0, 4000. 0, 6000. 0]" ); "lumi_kappa[1. 045]" ); "cexpr: : alpha_lumi('pow (lumi_kappa, beta_lumi)', lumi_kappa, beta_lumi[0, -5, 5])" ); p. Ws->factory( "prod: : lumi(lumi_nom, alpha_lumi)" ); p. Ws->factory( "Gaussian: : constr_lumi(beta_lumi, glob_lumi[0, -5, 5], 1)" ); Luca Lista Statistical Methods for Data Analysis 7

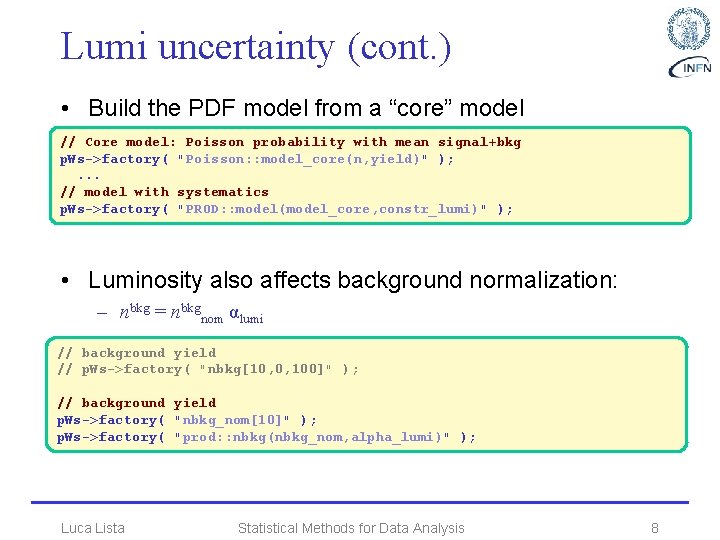
Lumi uncertainty (cont. ) • Build the PDF model from a “core” model // Core model: Poisson probability with mean signal+bkg p. Ws->factory( "Poisson: : model_core(n, yield)" ); . . . // model with systematics p. Ws->factory( "PROD: : model(model_core, constr_lumi)" ); • Luminosity also affects background normalization: – nbkg = nbkgnom αlumi // background yield // p. Ws->factory( "nbkg[10, 0, 100]" ); // background yield p. Ws->factory( "nbkg_nom[10]" ); p. Ws->factory( "prod: : nbkg(nbkg_nom, alpha_lumi)" ); Luca Lista Statistical Methods for Data Analysis 8

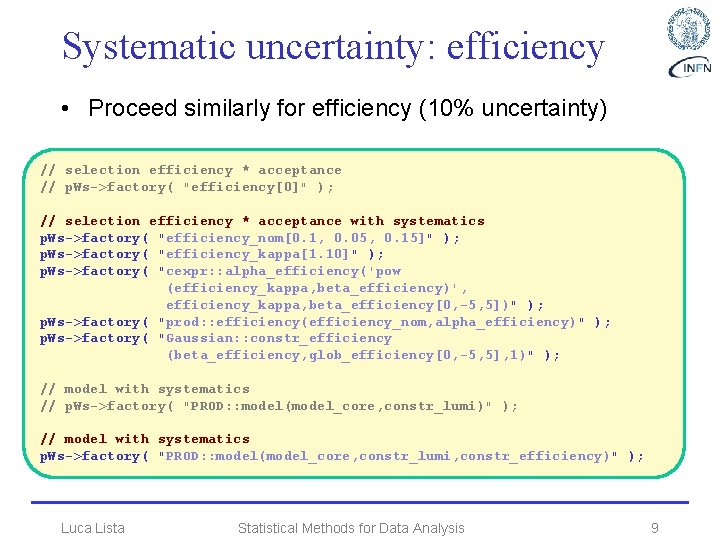
Systematic uncertainty: efficiency • Proceed similarly for efficiency (10% uncertainty) // selection efficiency * acceptance // p. Ws->factory( "efficiency[0]" ); // selection efficiency * acceptance with systematics p. Ws->factory( "efficiency_nom[0. 1, 0. 05, 0. 15]" ); p. Ws->factory( "efficiency_kappa[1. 10]" ); p. Ws->factory( "cexpr: : alpha_efficiency('pow (efficiency_kappa, beta_efficiency)', efficiency_kappa, beta_efficiency[0, -5, 5])" ); p. Ws->factory( "prod: : efficiency(efficiency_nom, alpha_efficiency)" ); p. Ws->factory( "Gaussian: : constr_efficiency (beta_efficiency, glob_efficiency[0, -5, 5], 1)" ); // model with systematics // p. Ws->factory( "PROD: : model(model_core, constr_lumi)" ); // model with systematics p. Ws->factory( "PROD: : model(model_core, constr_lumi, constr_efficiency)" ); Luca Lista Statistical Methods for Data Analysis 9

Systematic uncertainty: nbkg • Proceed similarly for nbkg (10% uncertainty) // background yield // p. Ws->factory( "nbkg_nom[10]" ); // background p. Ws->factory( yield with systematics "nbkg_nom[10. 0, 5. 0, 15. 0]" ); "nbkg_kappa[1. 10]" ); "cexpr: : alpha_nbkg('pow (nbkg_kappa, beta_nbkg)', nbkg_kappa, beta_nbkg[0, -5, 5])" ); p. Ws->factory( "prod: : nbkg(nbkg_nom, alpha_lumi, alpha_nbkg)" ); p. Ws->factory( "Gaussian: : constr_nbkg(beta_nbkg, glob_nbkg[0, -5, 5], 1)" ); // model with systematics // p. Ws->factory( "PROD: : model(model_core, constr_lumi, constr_efficiency)" ); // model with systematics p. Ws->factory( "PROD: : model (model_core, constr_lumi, constr_efficiency, constr_nbkg)" ); Luca Lista Statistical Methods for Data Analysis 10

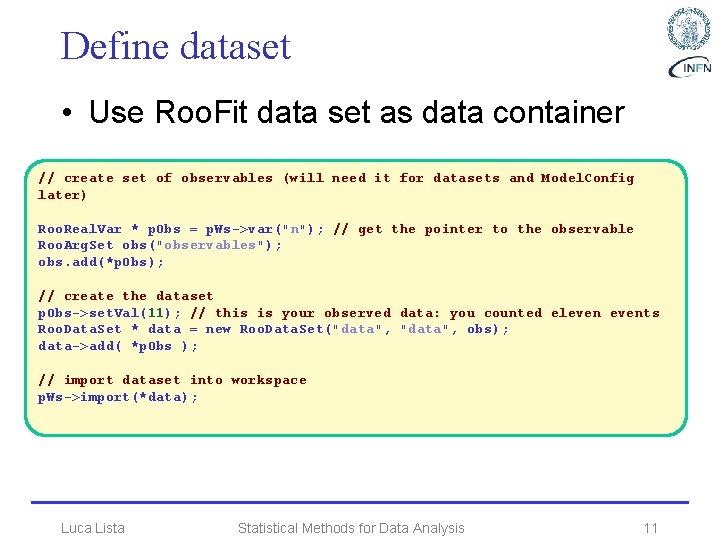
Define dataset • Use Roo. Fit data set as data container // create set of observables (will need it for datasets and Model. Config later) Roo. Real. Var * p. Obs = p. Ws->var("n"); // get the pointer to the observable Roo. Arg. Set obs("observables"); obs. add(*p. Obs); // create the dataset p. Obs->set. Val(11); // this is your observed data: you counted elevents Roo. Data. Set * data = new Roo. Data. Set("data", obs); data->add( *p. Obs ); // import dataset into workspace p. Ws->import(*data); Luca Lista Statistical Methods for Data Analysis 11

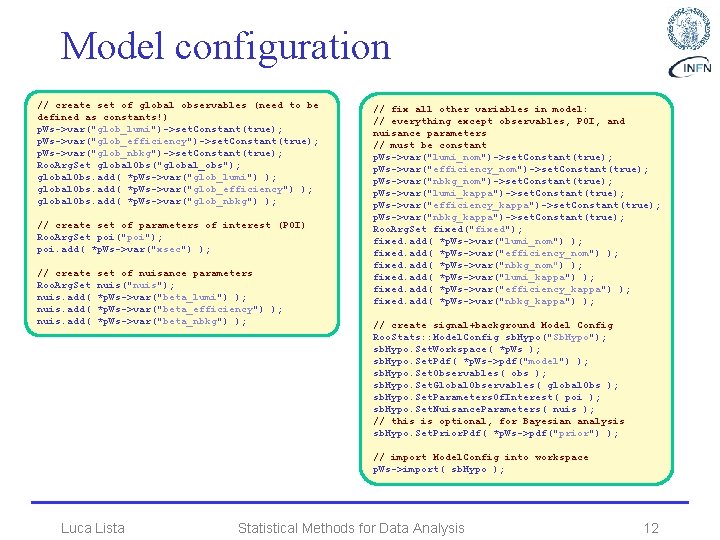
Model configuration // create set of global observables (need to be defined as constants!) p. Ws->var("glob_lumi")->set. Constant(true); p. Ws->var("glob_efficiency")->set. Constant(true); p. Ws->var("glob_nbkg")->set. Constant(true); Roo. Arg. Set global. Obs("global_obs"); global. Obs. add( *p. Ws->var("glob_lumi") ); global. Obs. add( *p. Ws->var("glob_efficiency") ); global. Obs. add( *p. Ws->var("glob_nbkg") ); // create set of parameters of interest (POI) Roo. Arg. Set poi("poi"); poi. add( *p. Ws->var("xsec") ); // create Roo. Arg. Set nuis. add( set of nuisance parameters nuis("nuis"); *p. Ws->var("beta_lumi") ); *p. Ws->var("beta_efficiency") ); *p. Ws->var("beta_nbkg") ); // fix all other variables in model: // everything except observables, POI, and nuisance parameters // must be constant p. Ws->var("lumi_nom")->set. Constant(true); p. Ws->var("efficiency_nom")->set. Constant(true); p. Ws->var("nbkg_nom")->set. Constant(true); p. Ws->var("lumi_kappa")->set. Constant(true); p. Ws->var("efficiency_kappa")->set. Constant(true); p. Ws->var("nbkg_kappa")->set. Constant(true); Roo. Arg. Set fixed("fixed"); fixed. add( *p. Ws->var("lumi_nom") ); fixed. add( *p. Ws->var("efficiency_nom") ); fixed. add( *p. Ws->var("nbkg_nom") ); fixed. add( *p. Ws->var("lumi_kappa") ); fixed. add( *p. Ws->var("efficiency_kappa") ); fixed. add( *p. Ws->var("nbkg_kappa") ); // create signal+background Model Config Roo. Stats: : Model. Config sb. Hypo("Sb. Hypo"); sb. Hypo. Set. Workspace( *p. Ws ); sb. Hypo. Set. Pdf( *p. Ws->pdf("model") ); sb. Hypo. Set. Observables( obs ); sb. Hypo. Set. Global. Observables( global. Obs ); sb. Hypo. Set. Parameters. Of. Interest( poi ); sb. Hypo. Set. Nuisance. Parameters( nuis ); // this is optional, for Bayesian analysis sb. Hypo. Set. Prior. Pdf( *p. Ws->pdf("prior") ); // import Model. Config into workspace p. Ws->import( sb. Hypo ); Luca Lista Statistical Methods for Data Analysis 12

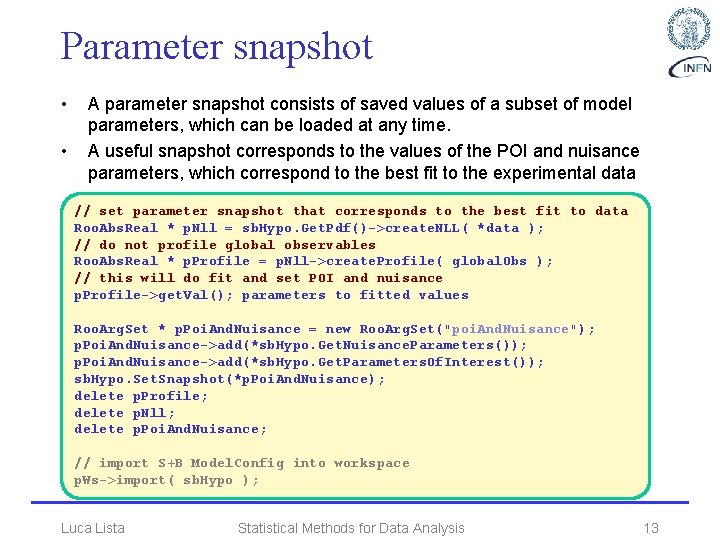
Parameter snapshot • • A parameter snapshot consists of saved values of a subset of model parameters, which can be loaded at any time. A useful snapshot corresponds to the values of the POI and nuisance parameters, which correspond to the best fit to the experimental data // set parameter snapshot that corresponds to the best fit to data Roo. Abs. Real * p. Nll = sb. Hypo. Get. Pdf()->create. NLL( *data ); // do not profile global observables Roo. Abs. Real * p. Profile = p. Nll->create. Profile( global. Obs ); // this will do fit and set POI and nuisance p. Profile->get. Val(); parameters to fitted values Roo. Arg. Set * p. Poi. And. Nuisance = new Roo. Arg. Set("poi. And. Nuisance"); p. Poi. And. Nuisance->add(*sb. Hypo. Get. Nuisance. Parameters()); p. Poi. And. Nuisance->add(*sb. Hypo. Get. Parameters. Of. Interest()); sb. Hypo. Set. Snapshot(*p. Poi. And. Nuisance); delete p. Profile; delete p. Nll; delete p. Poi. And. Nuisance; // import S+B Model. Config into workspace p. Ws->import( sb. Hypo ); Luca Lista Statistical Methods for Data Analysis 13

Adding more hypotheses models • More than one Model. Config can be added to the workspace for later use // create background-only Model Config from the S+B one Roo. Stats: : Model. Config b. Hypo = sb. Hypo; b. Hypo. Set. Name("BHypo"); b. Hypo. Set. Workspace(*p. Ws); // set parameter snapshot for b. Hypo, setting xsec=0 // it is useful to understand how this block of code works // but you can also use it as a recipe to make a parameter snapshot p. Nll = b. Hypo. Get. Pdf()->create. NLL( *data ); Roo. Arg. Set poi. And. Global. Obs("poi. And. Global. Obs"); poi. And. Global. Obs. add( poi ); poi. And. Global. Obs. add( global. Obs ); // do not profile POI and global observables p. Profile = p. Nll->create. Profile( poi. And. Global. Obs ); ((Roo. Real. Var *)poi. first())->set. Val( 0 ); // set xsec=0 here p. Profile->get. Val(); // this will do fit and set nuisance parameters to profiled values p. Poi. And. Nuisance = new Roo. Arg. Set( "poi. And. Nuisance" ); p. Poi. And. Nuisance->add( nuis ); p. Poi. And. Nuisance->add( poi ); b. Hypo. Set. Snapshot(*p. Poi. And. Nuisance); delete p. Profile; delete p. Nll; delete p. Poi. And. Nuisance; // import model config into workspace b. Hypo. Set. Workspace(*p. Ws); p. Ws->import( b. Hypo ); Luca Lista Statistical Methods for Data Analysis 14

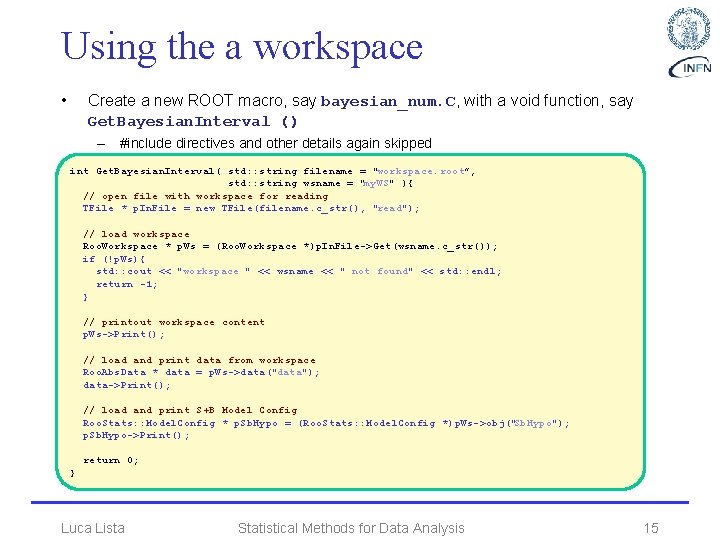
Using the a workspace • Create a new ROOT macro, say bayesian_num. C, with a void function, say Get. Bayesian. Interval () – #include directives and other details again skipped int Get. Bayesian. Interval( std: : string filename = "workspace. root”, std: : string wsname = "my. WS" ){ // open file with workspace for reading TFile * p. In. File = new TFile(filename. c_str(), "read"); // load workspace Roo. Workspace * p. Ws = (Roo. Workspace *)p. In. File->Get(wsname. c_str()); if (!p. Ws){ std: : cout << "workspace " << wsname << " not found" << std: : endl; return -1; } // printout workspace content p. Ws->Print(); // load and print data from workspace Roo. Abs. Data * data = p. Ws->data("data"); data->Print(); // load and print S+B Model Config Roo. Stats: : Model. Config * p. Sb. Hypo = (Roo. Stats: : Model. Config *)p. Ws->obj("Sb. Hypo"); p. Sb. Hypo->Print(); return 0; } Luca Lista Statistical Methods for Data Analysis 15

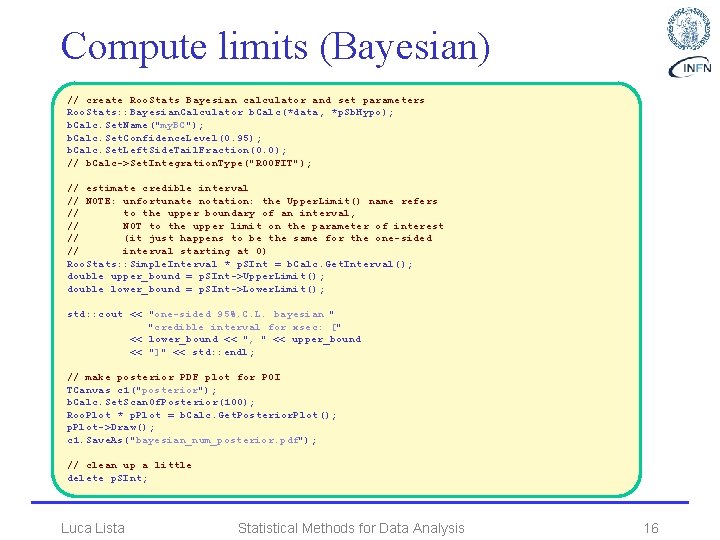
Compute limits (Bayesian) // create Roo. Stats Bayesian calculator and set parameters Roo. Stats: : Bayesian. Calculator b. Calc(*data, *p. Sb. Hypo); b. Calc. Set. Name("my. BC"); b. Calc. Set. Confidence. Level(0. 95); b. Calc. Set. Left. Side. Tail. Fraction(0. 0); // b. Calc->Set. Integration. Type("ROOFIT"); // estimate credible interval // NOTE: unfortunate notation: the Upper. Limit() name refers // to the upper boundary of an interval, // NOT to the upper limit on the parameter of interest // (it just happens to be the same for the one-sided // interval starting at 0) Roo. Stats: : Simple. Interval * p. SInt = b. Calc. Get. Interval(); double upper_bound = p. SInt->Upper. Limit(); double lower_bound = p. SInt->Lower. Limit(); std: : cout << "one-sided 95%. C. L. bayesian " "credible interval for xsec: [" << lower_bound << ", " << upper_bound << "]" << std: : endl; // make posterior PDF plot for POI TCanvas c 1("posterior"); b. Calc. Set. Scan. Of. Posterior(100); Roo. Plot * p. Plot = b. Calc. Get. Posterior. Plot(); p. Plot->Draw(); c 1. Save. As("bayesian_num_posterior. pdf"); // clean up a little delete p. SInt; Luca Lista Statistical Methods for Data Analysis 16

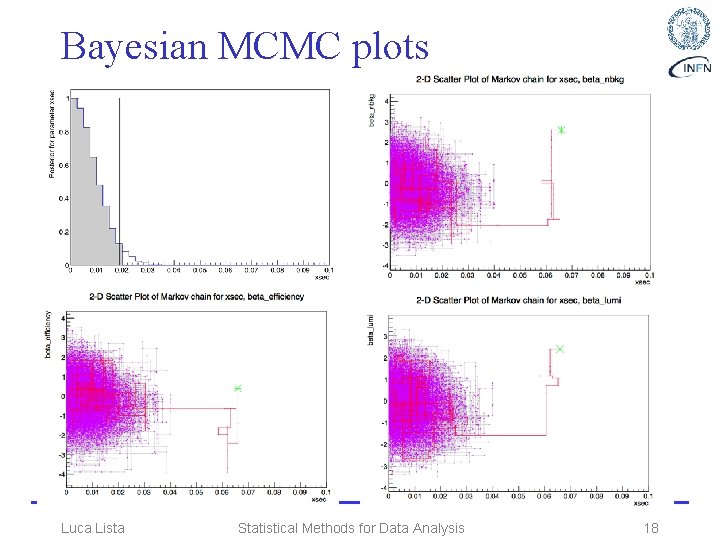
Bayesian Markov Chain • Copy bayesian_num. C to bayesian_mcmc. C and insert the code below // Metropolis-Hastings algorithm needs a proposal function Roo. Stats: : Sequential. Proposal sp(10. 0); Roo. Stats: : MCMCCalculator mcmc( *data, *p. Sb. Hypo ); mcmc. Set. Confidence. Level(0. 95); mcmc. Set. Num. Iters(100000); //num. iterations mcmc. Set. Proposal. Function(sp); //first N steps to be ignored as burn-in mcmc. Set. Num. Burn. In. Steps(500); mcmc. Set. Left. Side. Tail. Fraction(0. 0); //binning for plotting only mcmc. Set. Num. Bins(40); // estimate credible interval Roo. Stats: : MCMCInterval * p. Mcmc. Int = mcmc. Get. Interval(); double upper_bound = p. Mcmc. Int->Upper. Limit( *p. Ws->var("xsec") ); double lower_bound = p. Mcmc. Int->Lower. Limit( *p. Ws->var("xsec") ); std: : cout << "one-sided 95%. C. L. bayesian " " credible interval for xsec: [" << lower_bound << ", " << upper_bound << "]" << std: : endl; Luca Lista // make posterior PDF plot for POI TCanvas c 1("posterior"); Roo. Stats: : MCMCInterval. Plot plot(*p. Mcmc. Int); plot. Draw(); c 1. Save. As("bayesian_mcmc_posterior. pdf"); // make scatter plots to visualise the Marov chain TCanvas c 2("xsec_vs_beta_lumi"); plot. Draw. Chain. Scatter( *p. Ws->var("xsec"), *p. Ws->var("beta_lumi")); c 2. Save. As("scatter_mcmc_xsec_vs_beta_lumi. pdf"); TCanvas c 3("xsec_vs_beta_efficiency"); plot. Draw. Chain. Scatter( *p. Ws->var("xsec"), *p. Ws->var("beta_efficiency")); c 3. Save. As("scatter_xsec_vs_beta_efficiency. pdf"); TCanvas c 4("xsec_vs_beta_nbkg"); plot. Draw. Chain. Scatter( *p. Ws->var("xsec"), *p. Ws->var("beta_nbkg")); c 4. Save. As("scatter_xsec_vs_beta_nbkg. pdf"); // clean up a little delete p. Mcmc. Int; Statistical Methods for Data Analysis 17

Bayesian MCMC plots Luca Lista Statistical Methods for Data Analysis 18