Statistical Methods for Ancient Genomics 1 Thank You


















- Slides: 51

Statistical Methods for Ancient Genomics 1

Thank You • • Steven Senturia Bill Blumberg Linda Neumann Dan Mc. Carthy 2

Two Broad Categories of Methods Conventional Statistics • Principal Components Analysis • Cluster Analysis F Statistics • Genomic Distance • Three-Population Test • Four-Population Test 3

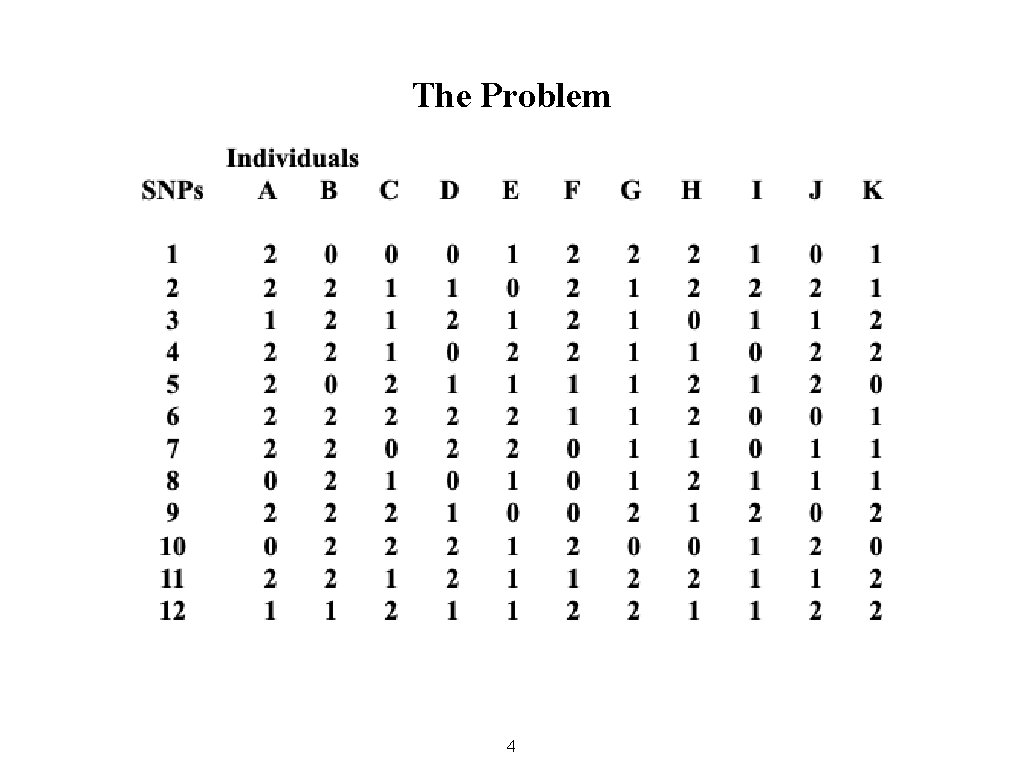
The Problem 4

Two Questions • Which people have a similar mix of SNPs? • Which SNPs have a similar mix of people? 5

Principal Components Analysis Analogies • Rotating a cube • Linear regression 6

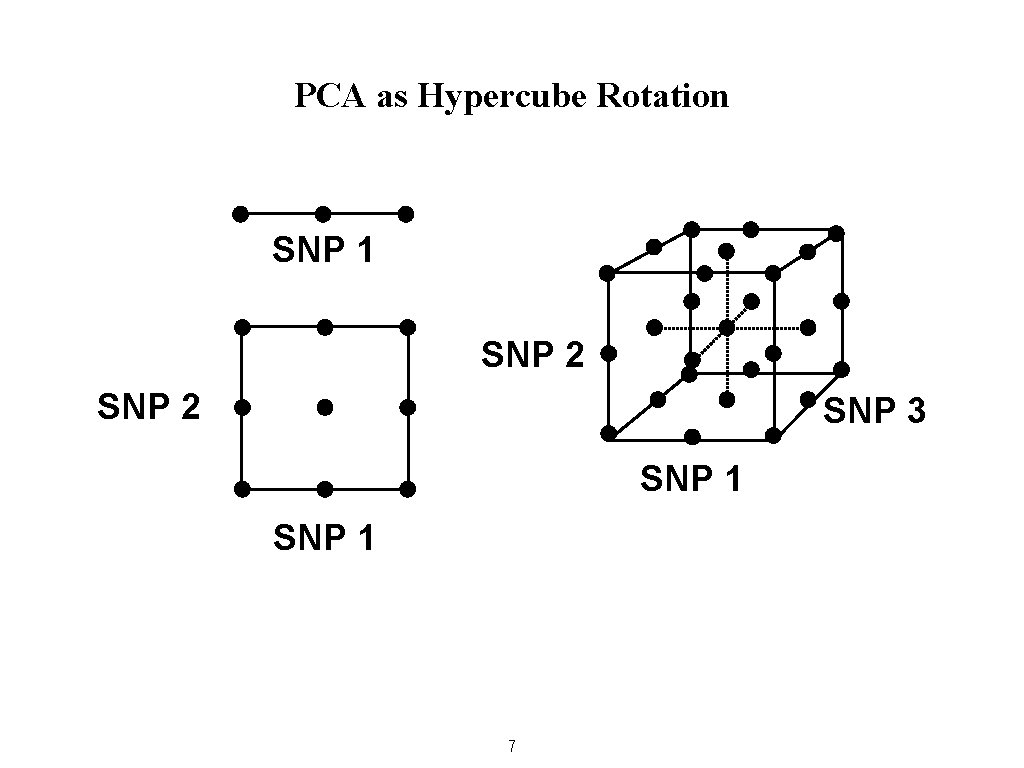
PCA as Hypercube Rotation SNP 1 SNP 2 SNP 3 SNP 1 7

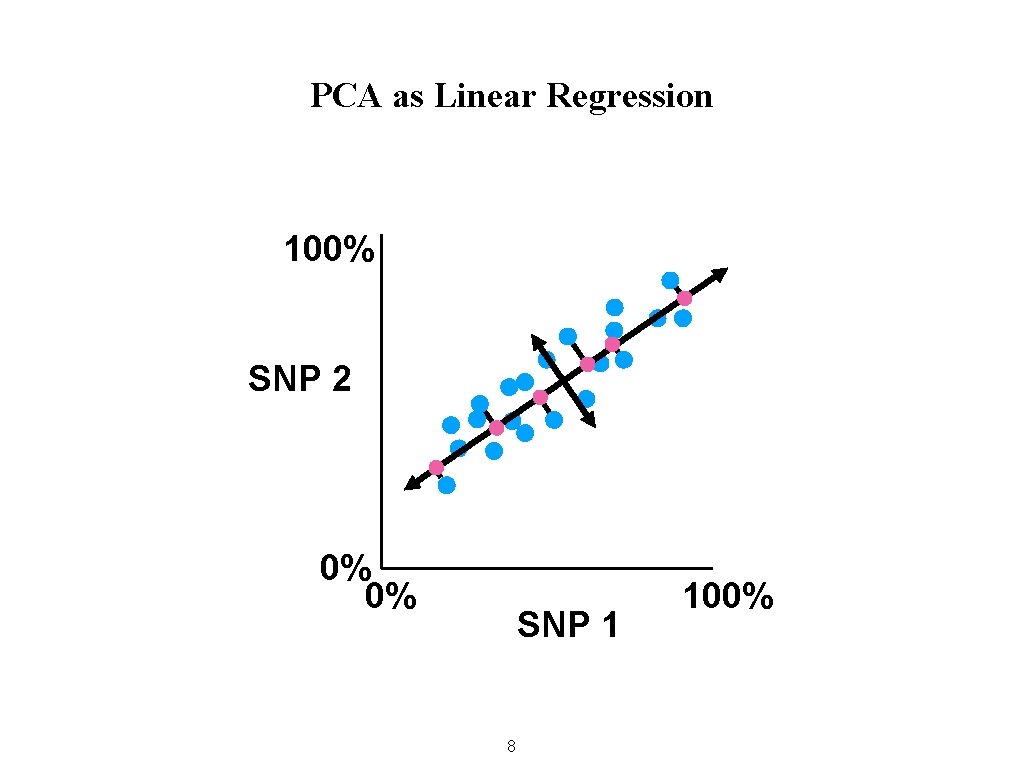
PCA as Linear Regression 100% SNP 2 0% 0% SNP 1 8 100%

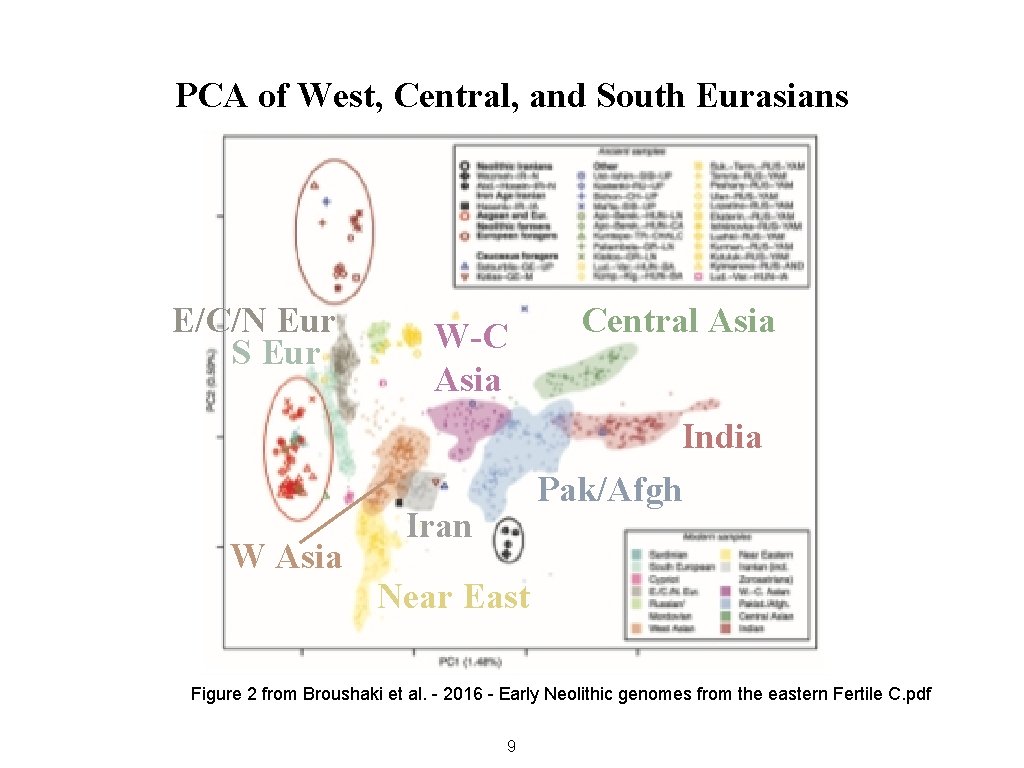
PCA of West, Central, and South Eurasians E/C/N Eur S Eur W Asia W-C Asia Central Asia India Pak/Afgh Iran Near East Figure 2 from Broushaki et al. - 2016 - Early Neolithic genomes from the eastern Fertile C. pdf 9

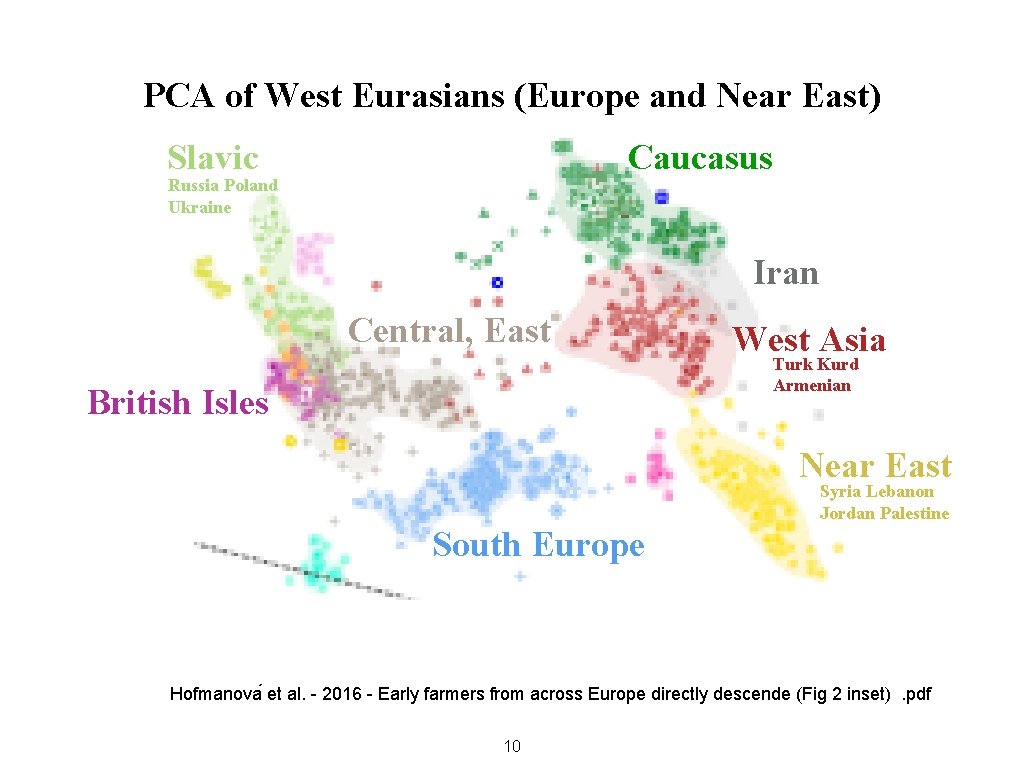
PCA of West Eurasians (Europe and Near East) Slavic Caucasus Russia Poland Ukraine Iran Central, East West Asia Turk Kurd Armenian British Isles Near East Syria Lebanon Jordan Palestine South Europe Hofmanova et al. - 2016 - Early farmers from across Europe directly descende (Fig 2 inset). pdf 10

Cluster Analysis Actually called • ADMIXTURE • STRUCTURE 11

Cluster Analysis Given the frequencies of many SNPs for many individuals. . . • How many ancestral populations contributed to these individuals? • What mix of SNPs did each ancestral population have? • What mix of ancestral populations does each individual have? 12

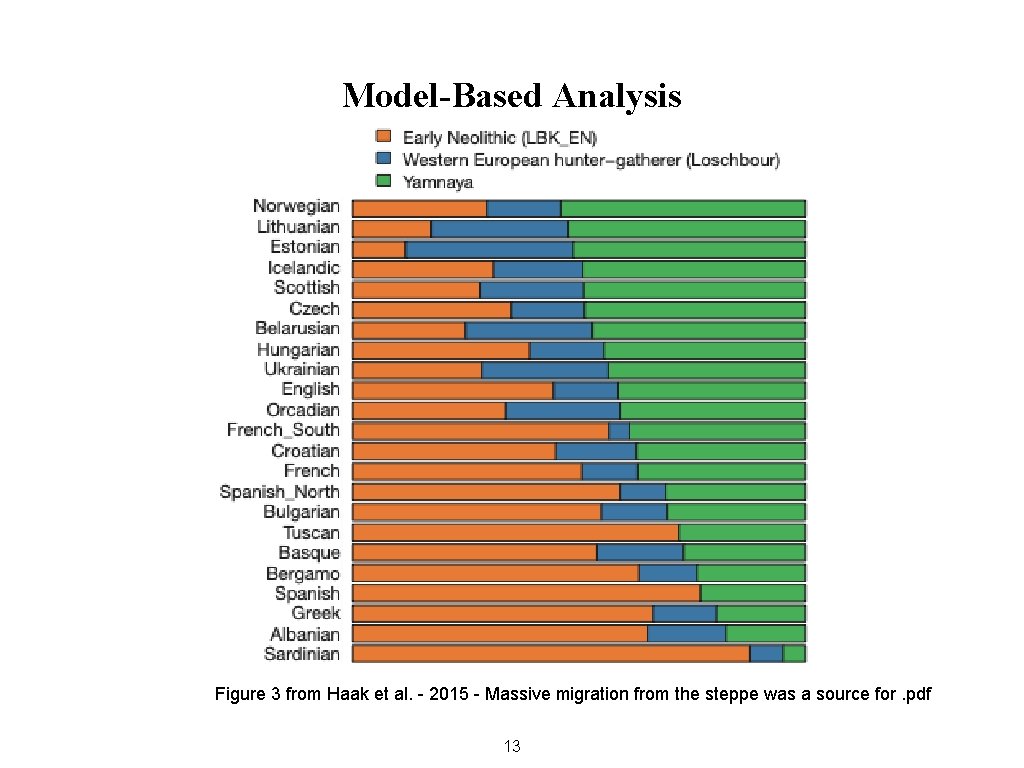
Model-Based Analysis Figure 3 from Haak et al. - 2015 - Massive migration from the steppe was a source for. pdf 13

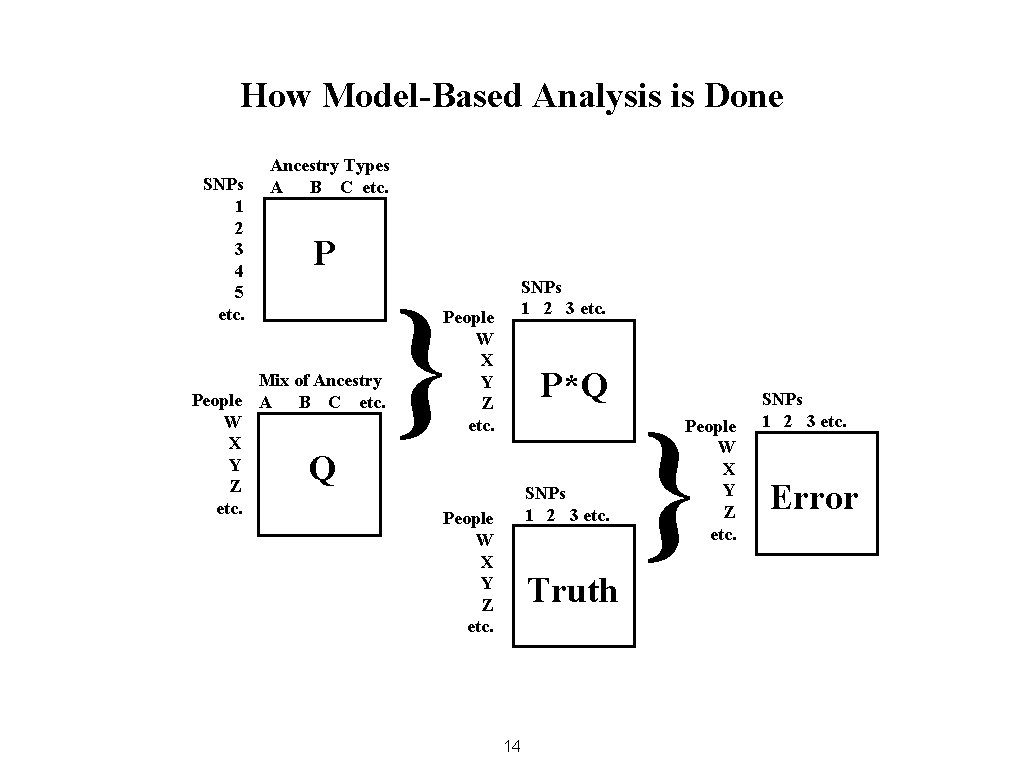
How Model-Based Analysis is Done SNPs 1 2 3 4 5 etc. Ancestry Types A B C etc. P Mix of Ancestry People A B C etc. W X Y Z etc. } SNPs 1 2 3 etc. People W X Y Z etc. P*Q Q SNPs 1 2 3 etc. People W X Y Z etc. Truth 14 } People W X Y Z etc. SNPs 1 2 3 etc. Error

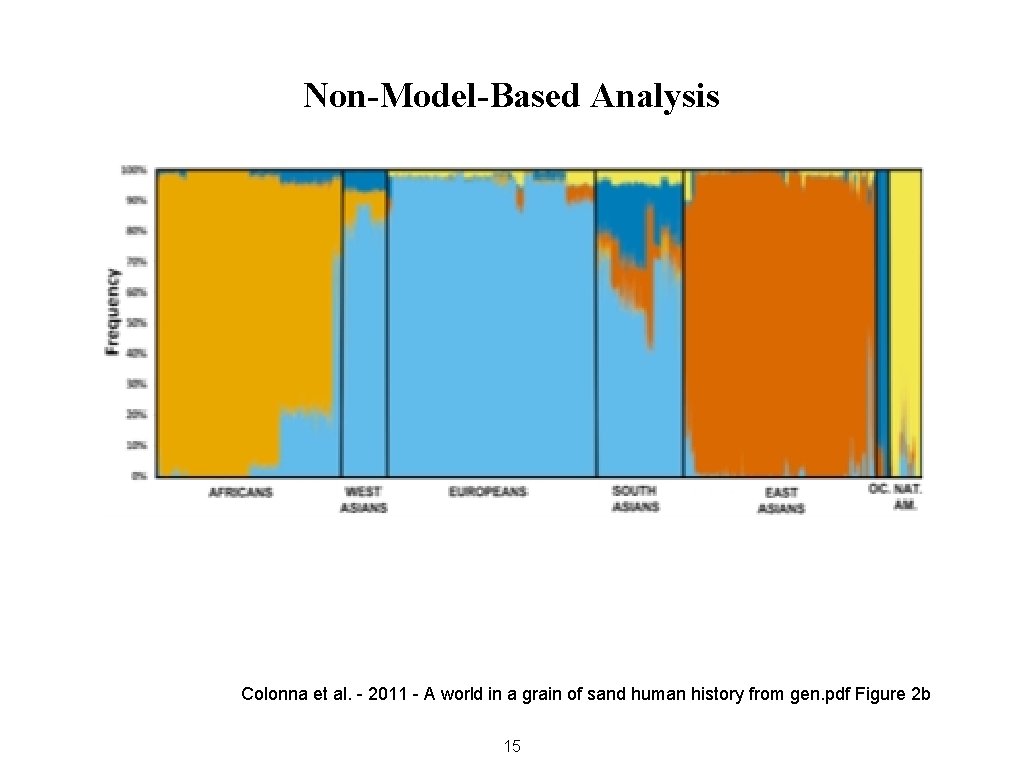
Non-Model-Based Analysis Colonna et al. - 2011 - A world in a grain of sand human history from gen. pdf Figure 2 b 15

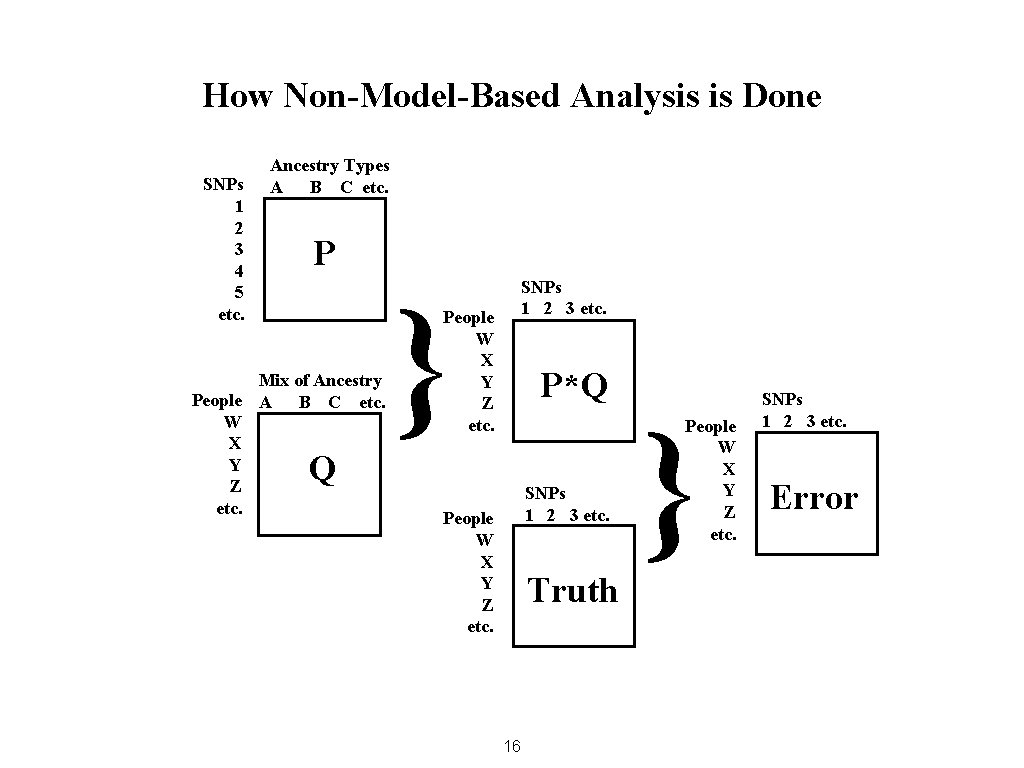
How Non-Model-Based Analysis is Done SNPs 1 2 3 4 5 etc. Ancestry Types A B C etc. P Mix of Ancestry People A B C etc. W X Y Z etc. } SNPs 1 2 3 etc. People W X Y Z etc. P*Q Q SNPs 1 2 3 etc. People W X Y Z etc. Truth 16 } People W X Y Z etc. SNPs 1 2 3 etc. Error

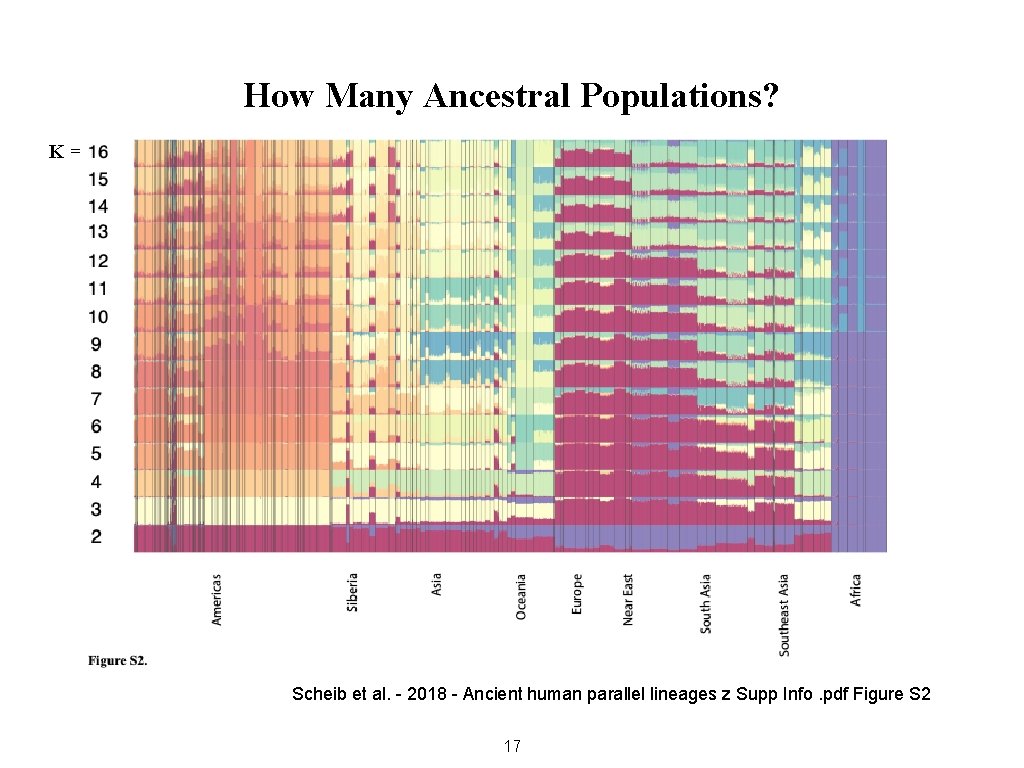
How Many Ancestral Populations? K= Scheib et al. - 2018 - Ancient human parallel lineages z Supp Info. pdf Figure S 2 17

Statistical Measures for Ancient Genomics Conventional Statistics • Principal Components Analysis • ADMIXTURE and STRUCTURE F Statistics • Genomic Distance • Three-Population Test • Four-Population Test 18

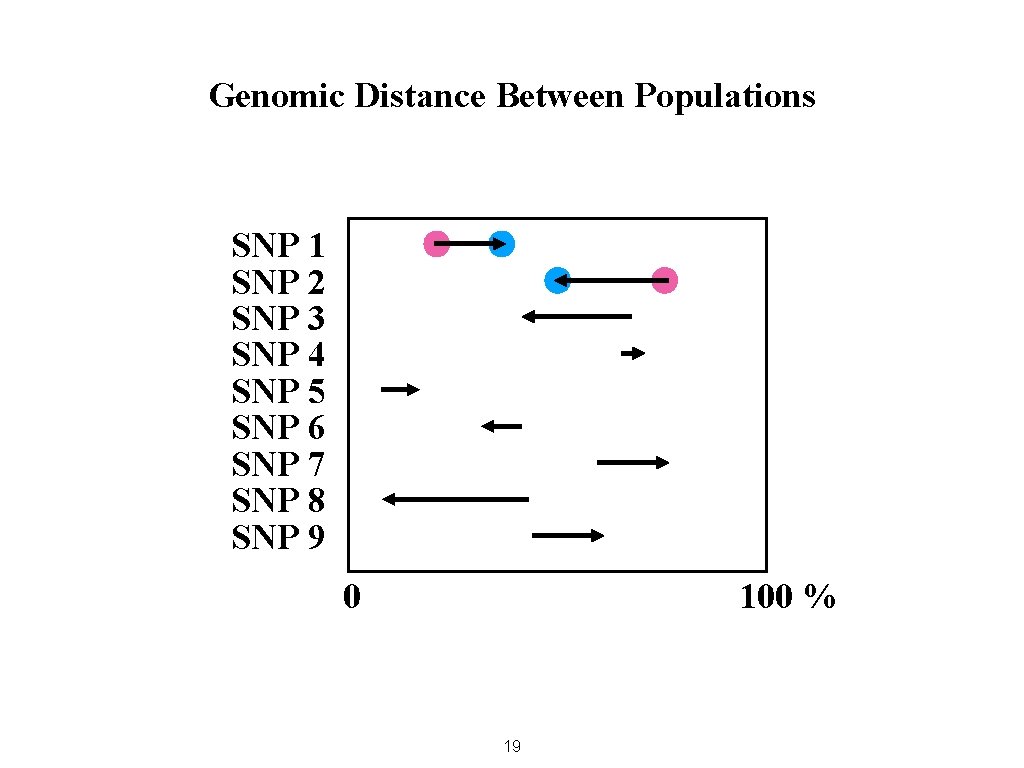
Genomic Distance Between Populations SNP 1 SNP 2 SNP 3 SNP 4 SNP 5 SNP 6 SNP 7 SNP 8 SNP 9 0 100 % 19

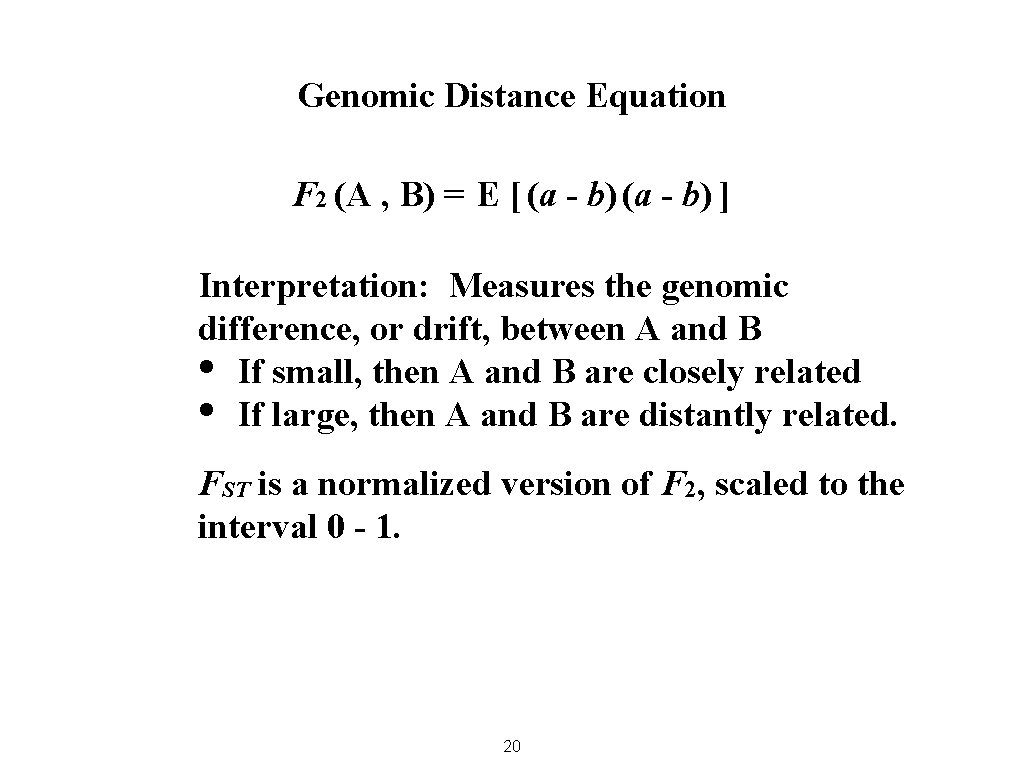
Genomic Distance Equation F 2 (A , B) = E [ (a - b) ] Interpretation: Measures the genomic difference, or drift, between A and B • If small, then A and B are closely related • If large, then A and B are distantly related. FST is a normalized version of F 2, scaled to the interval 0 - 1. 20

The Three-Population Test F 3 (A , B ; C) 21

Two SNPs over Time in a Population 100% SNP 1 Pop X SNP 2 0% Time 22

One SNP over Time in Two Populations 100% Pop 1 SNP X Pop 2 0% Time 23

The Three-Population Test Equation F 3 (A , B ; C) = E [ (c - a) (c - b) ] 24

100% SNP X Pop. 1 Pop. 2 0% Time 25

100% SNP X Pop. 1 Pop. 2 0% Time 26

100% Pop. 2 SNP X Pop. 1 0% Time 27

100% SNP X 0% Time 28

100% SNP X 0% Time 29

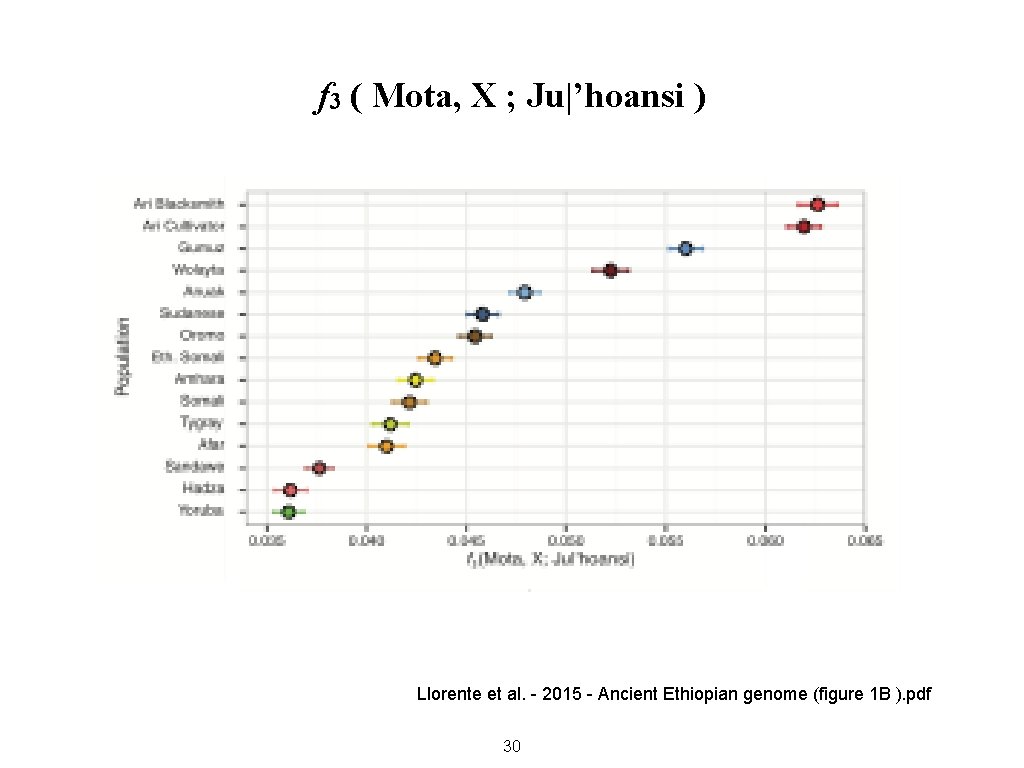
f 3 ( Mota, X ; Ju|’hoansi ) Llorente et al. - 2015 - Ancient Ethiopian genome (figure 1 B ). pdf 30

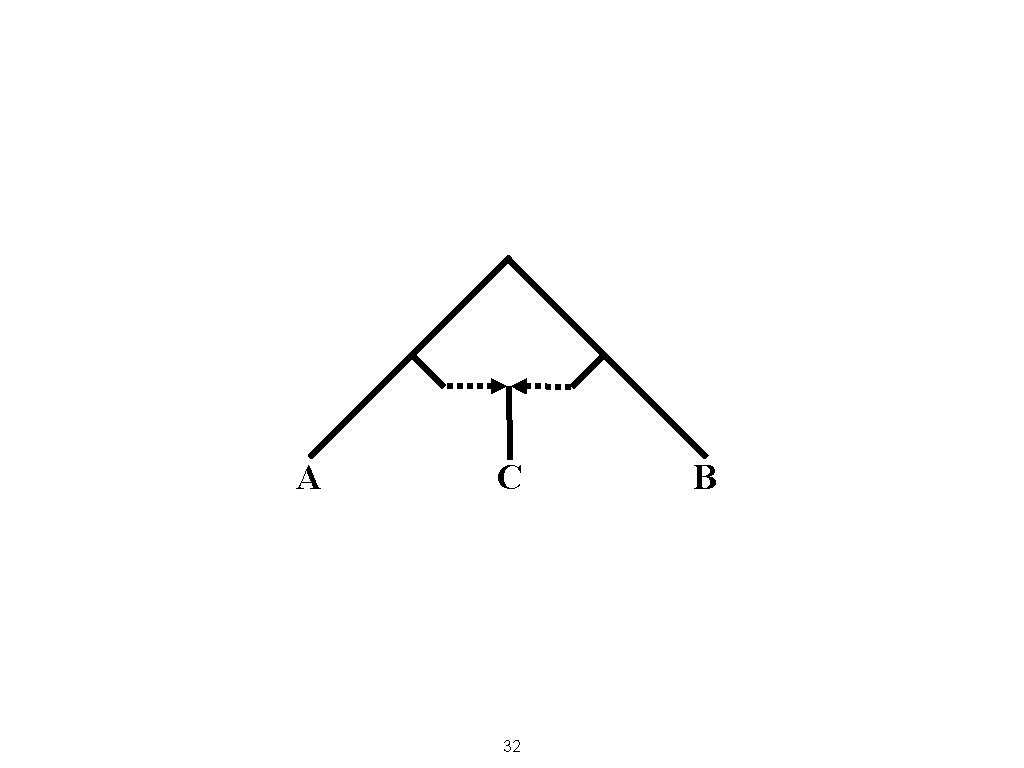
How could you get a negative f 3 result? 31

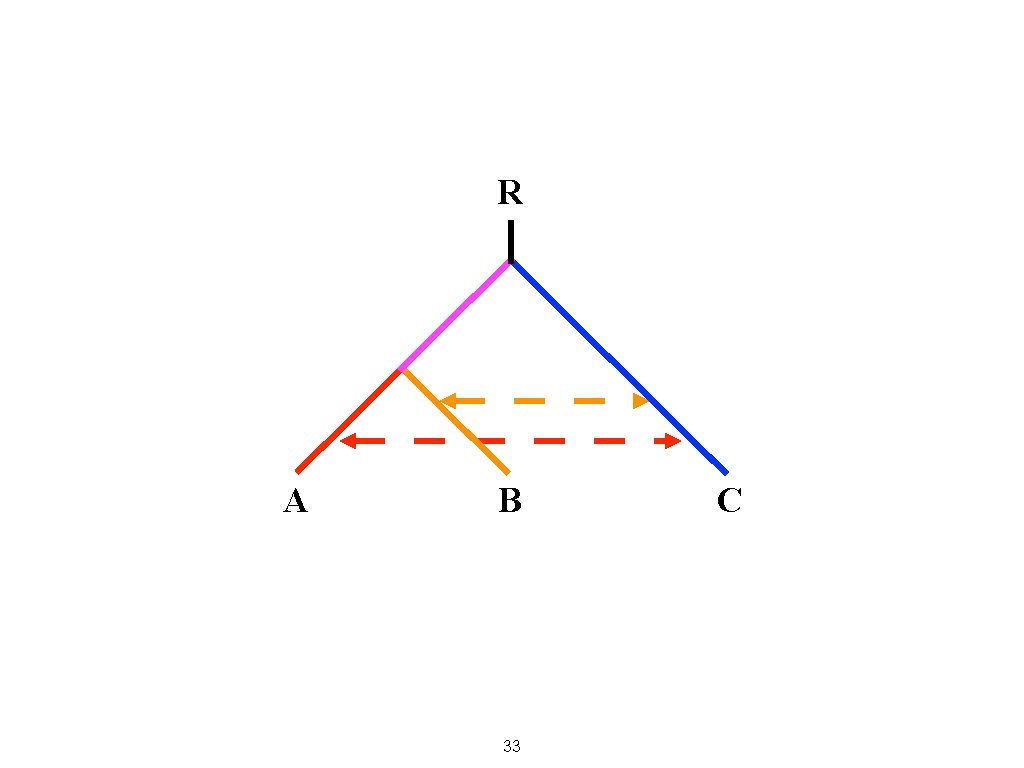
A C 32 B

R A B 33 C

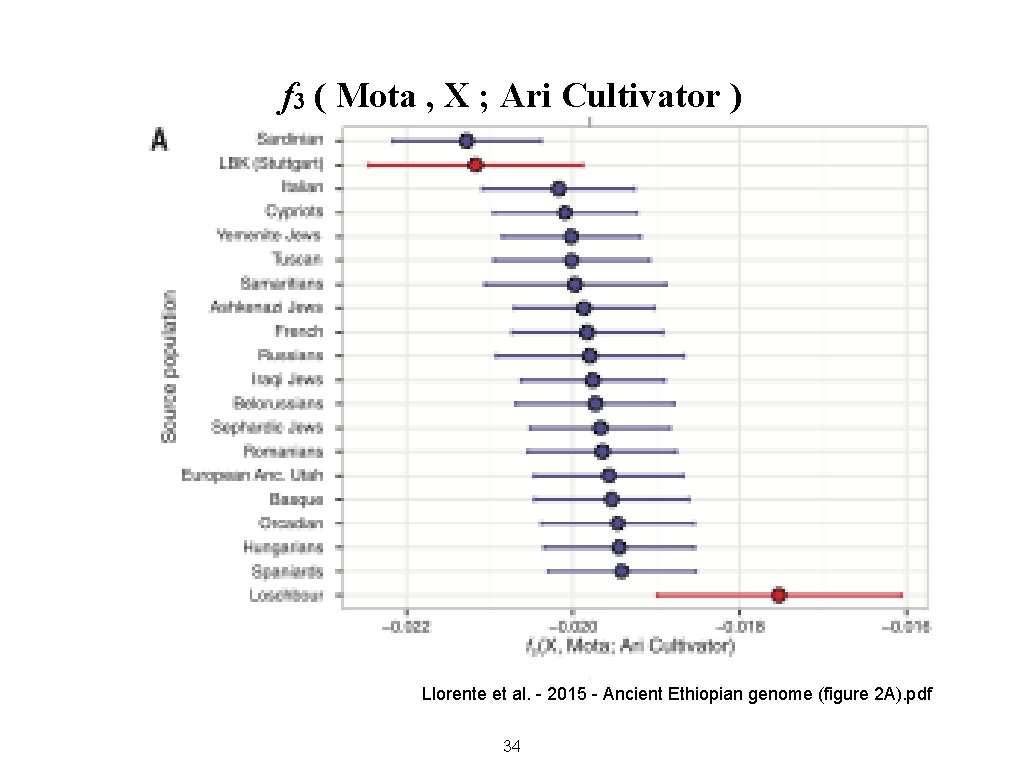
f 3 ( Mota , X ; Ari Cultivator ) Llorente et al. - 2015 - Ancient Ethiopian genome (figure 2 A). pdf 34

F 3 (A , B ; C) = E [ (c - a)(c - b) ] Interpretation: • If not significantly different from zero, the three populations are unrelated • If negative, then C is related to A and/or B, by admixture or common ancestry • If positive and if C is an outgroup, then the larger the result, the longer the shared drift of A and B the more closely related are A and B 35

The Four-Population Test F 4 (A , B ; C , D) = E [ (a - b)(c - d) ] 36
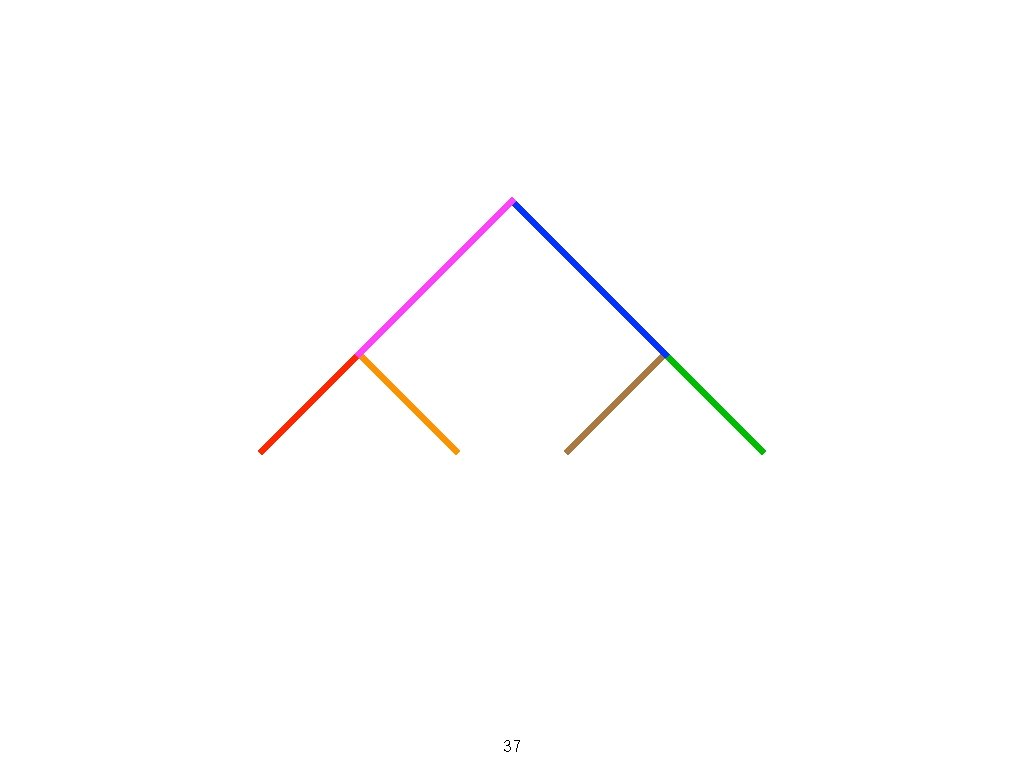
37

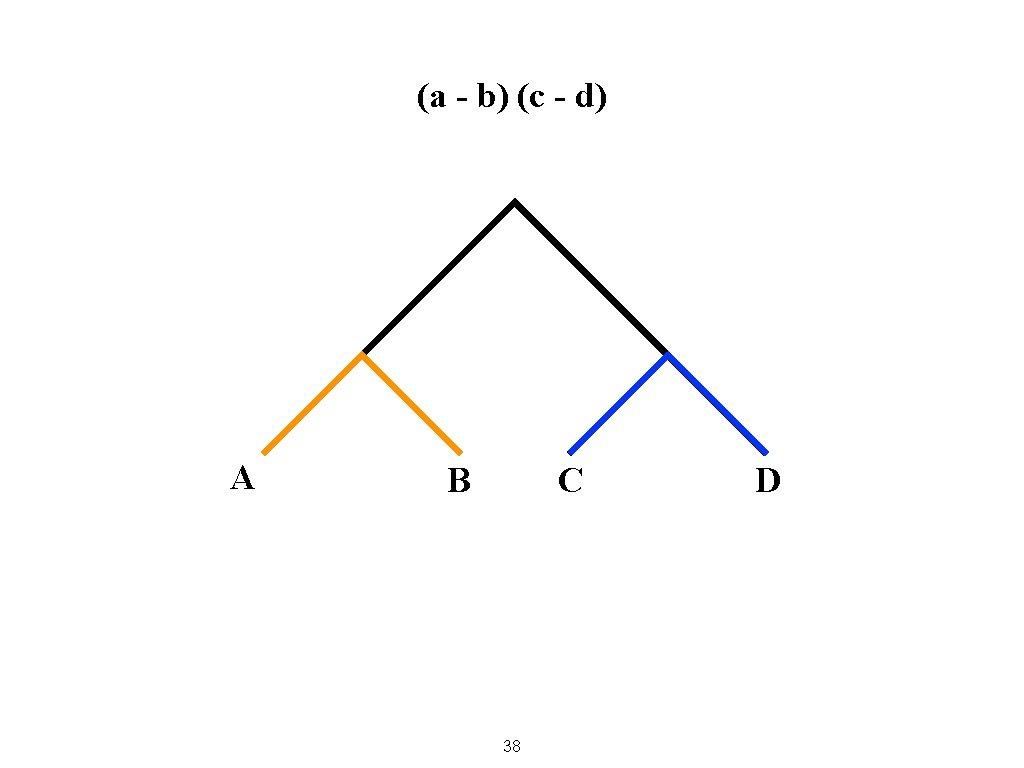
(a - b) (c - d) A B C 38 D

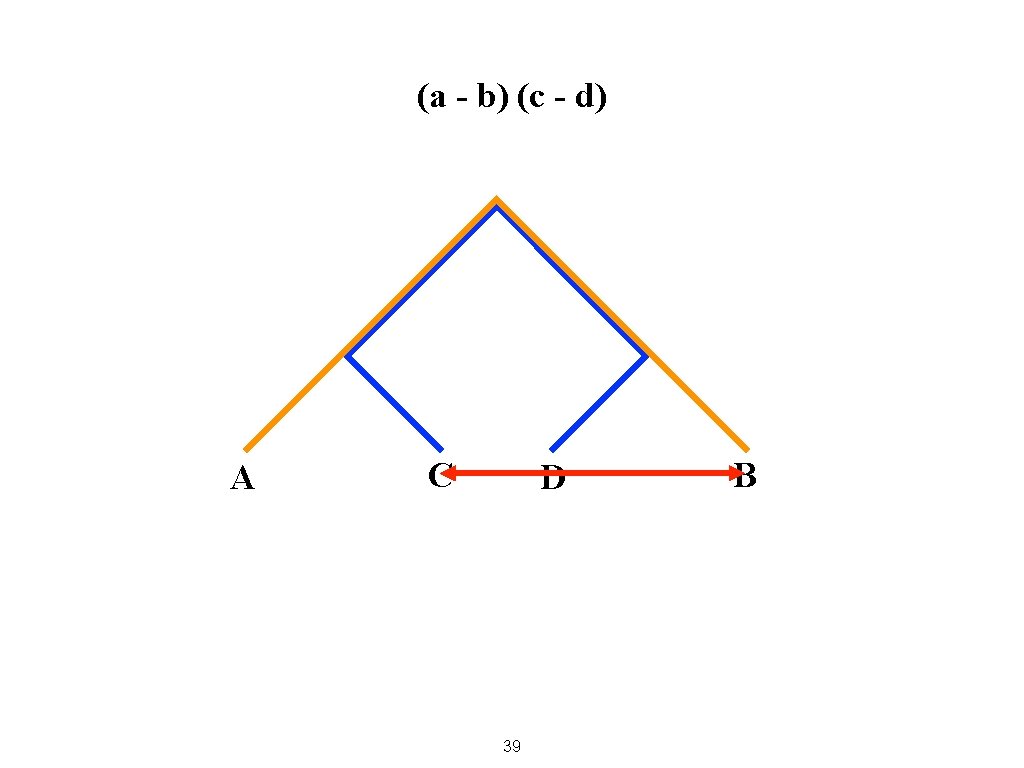
(a - b) (c - d) A C D 39 B

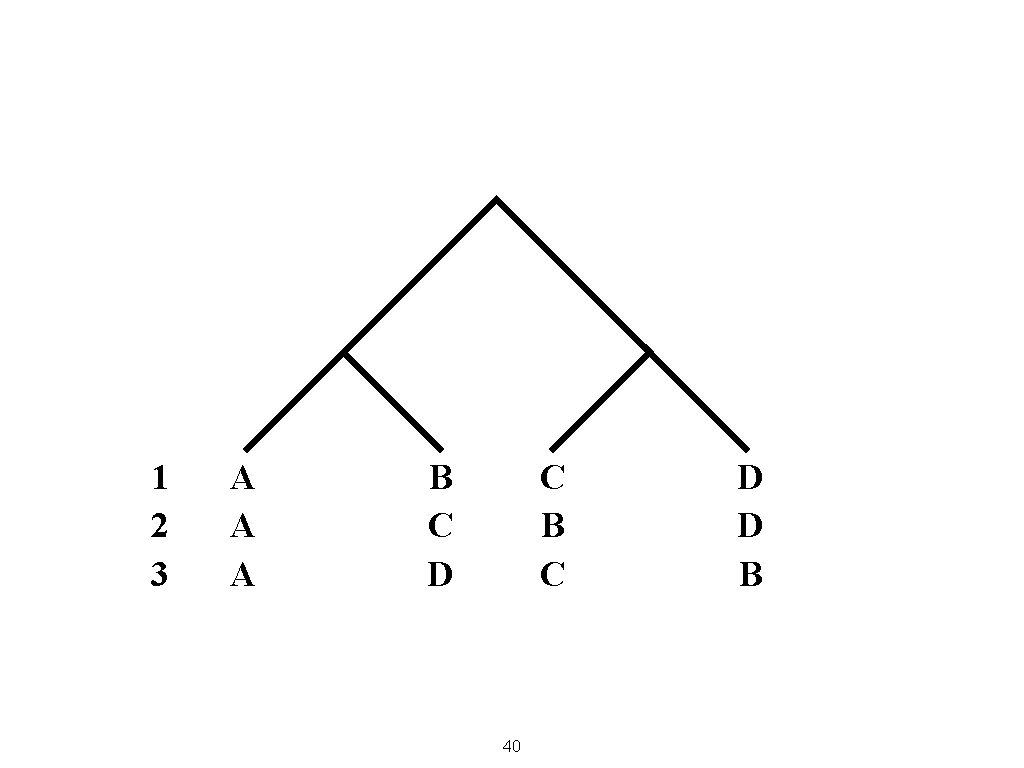
1 2 3 A A A B C D C B C 40 D D B

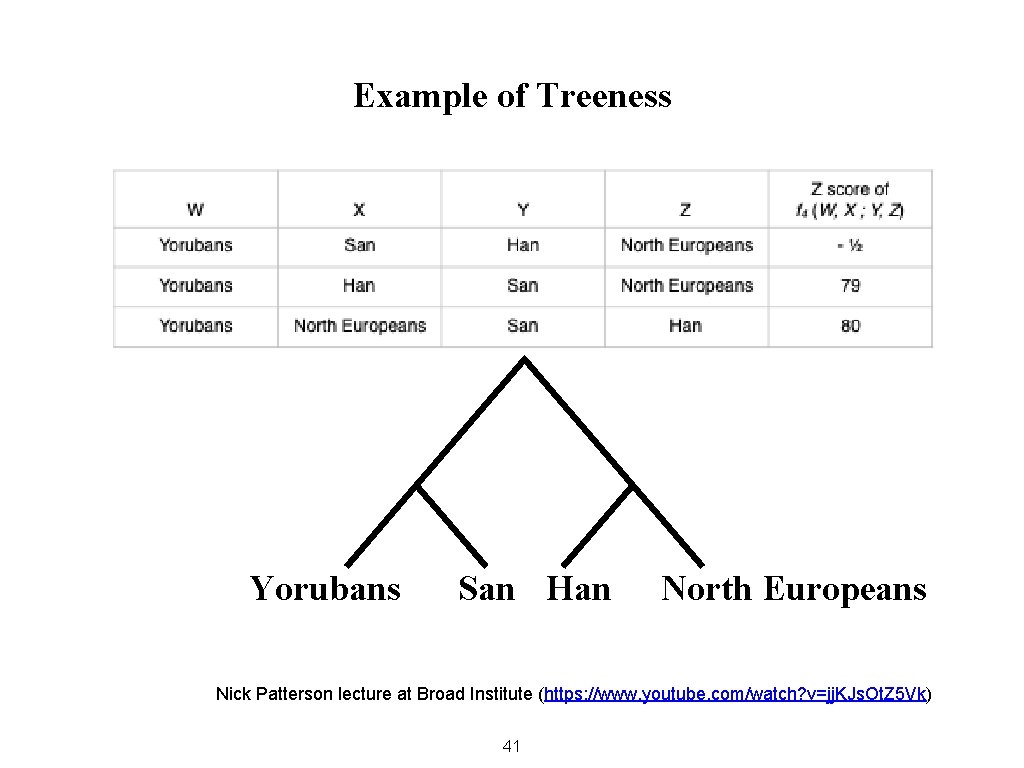
Example of Treeness Yorubans San Han North Europeans Nick Patterson lecture at Broad Institute (https: //www. youtube. com/watch? v=jj. KJs. Ot. Z 5 Vk) 41

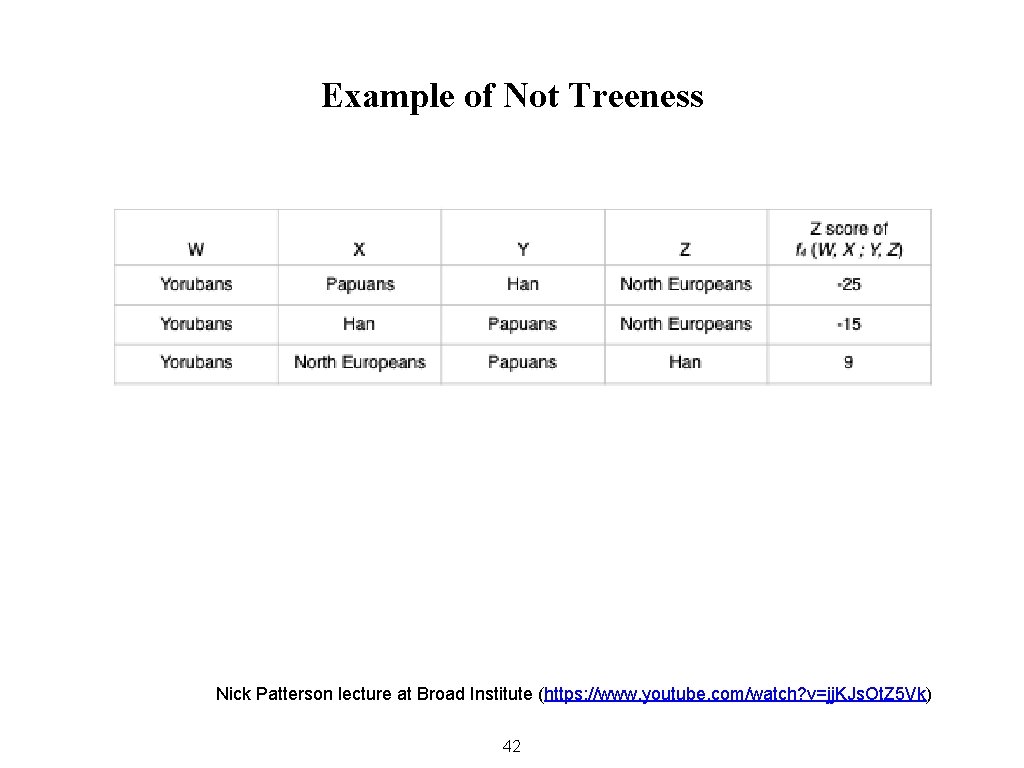
Example of Not Treeness Nick Patterson lecture at Broad Institute (https: //www. youtube. com/watch? v=jj. KJs. Ot. Z 5 Vk) 42

Uses of the Four-Population Test The first use of F 4 is to determine whether the relations between four populations can be accurately represented by a simple branching tree. 43

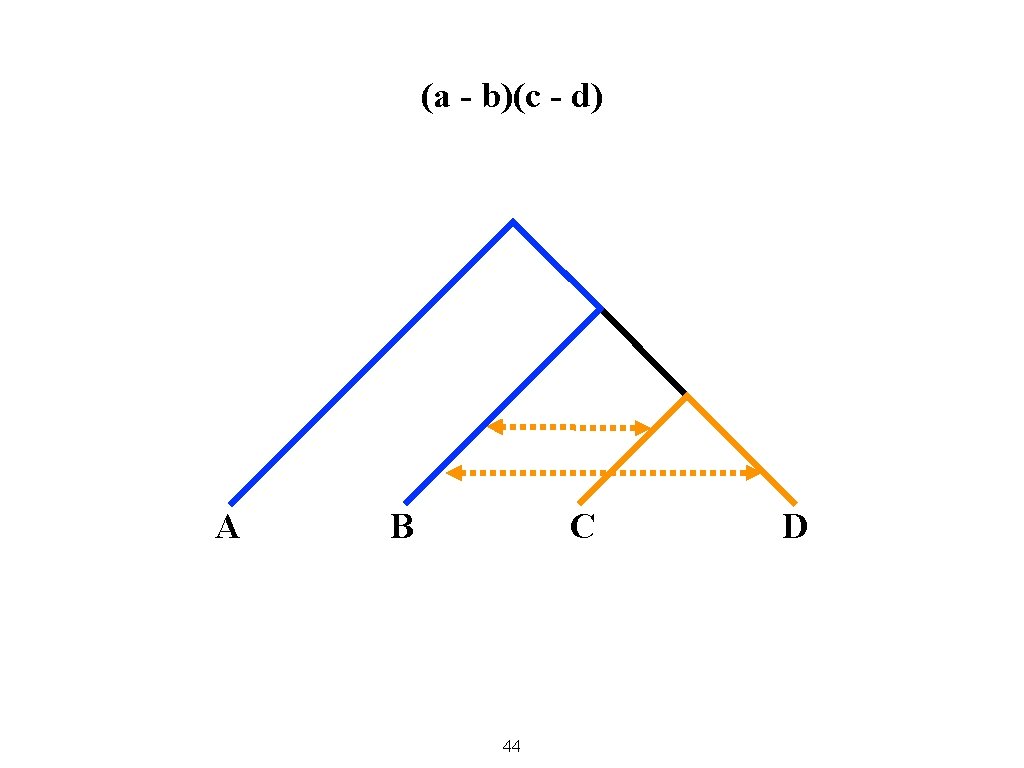
(a - b)(c - d) A B C 44 D

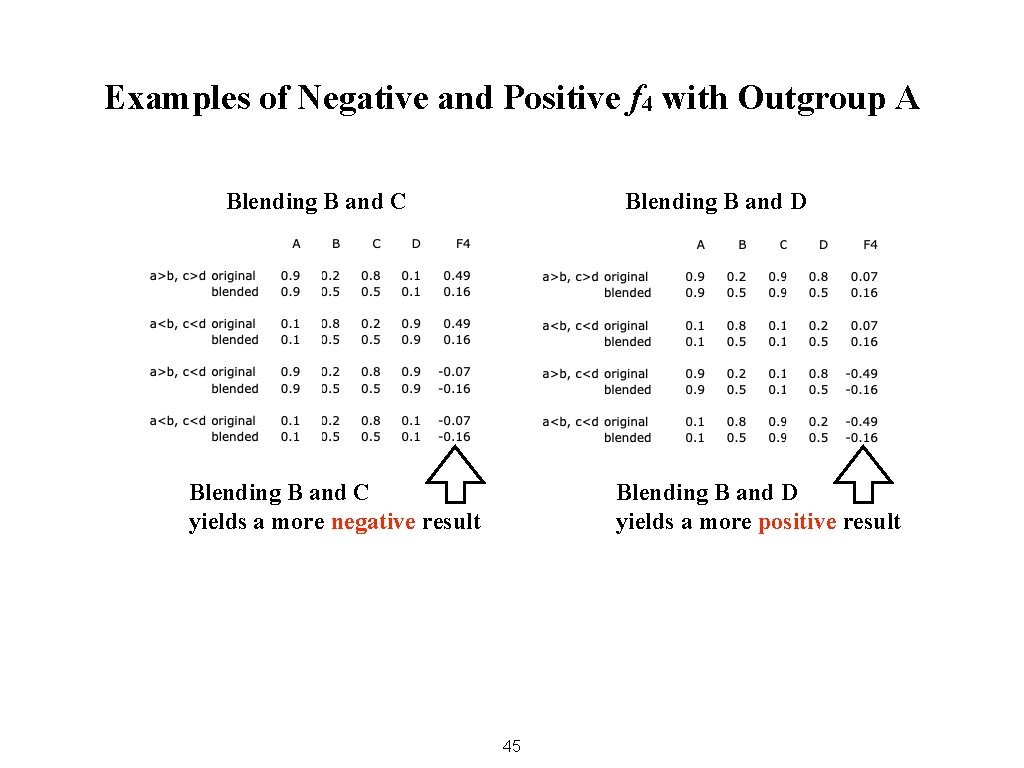
Examples of Negative and Positive f 4 with Outgroup A Blending B and C Blending B and D Blending B and C yields a more negative result Blending B and D yields a more positive result 45

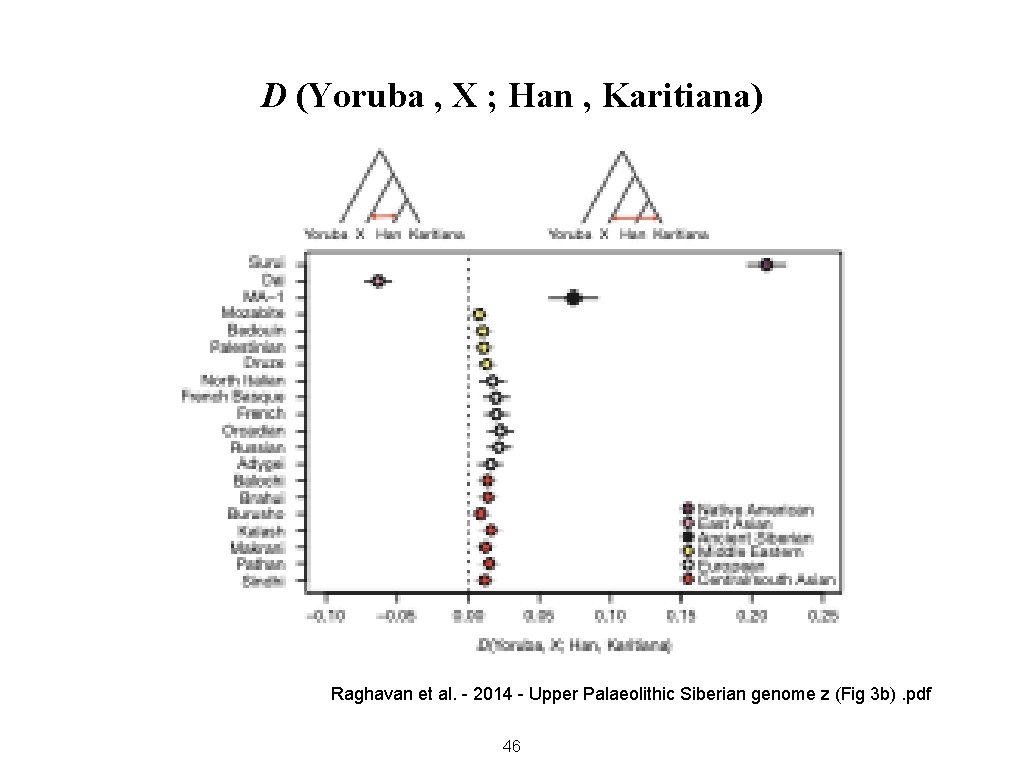
D (Yoruba , X ; Han , Karitiana) Raghavan et al. - 2014 - Upper Palaeolithic Siberian genome z (Fig 3 b). pdf 46

Uses of f 4 When population A is an outgroup, • A positive result means B is more closely related to D than to C • A negative result means B is more closely related to C than to D 47

What Would This Result Tell Us? f 4(Chimp, Neanderthal ; European, African) = -0. 2 if the standard error is 0. 01 The numbers are fictitious 48

Uses of f 4 Comparisons of relations between multiple F 4 tests on overlapping sets of populations can also be used to: • Determine the direction of gene flow in an admixture event • Measure the fraction of the ancestry of a “hybrid” population that came from each parent population 49

Interpreting f Statistics Handout 50

The End 51