Statistical Genomics Lecture 6 Genotyping By Sequencing Zhiwu

Statistical Genomics Lecture 6: Genotyping By Sequencing Zhiwu Zhang Washington State University

Outline • Genetic markers • Sequencing • Full vs. reduced

Human genome project • Funded by DOE, NIH and Welcome Trust in the UK • Begun in 1990 • Original planed to last 15 years. • Institute for Genomic Research and U. of Washington provided over 450 K BAC each was tagged and contain 3~4 Kb across the entire human genome

Human genome project • Accelerate the completion date to 2003 • Celera Genomics (shotgun approach) • Craig Venter was among those sequenced • Identified 20~120 K genes • Sequence of 3 billion base pairs • Cost near 3 billion dollars
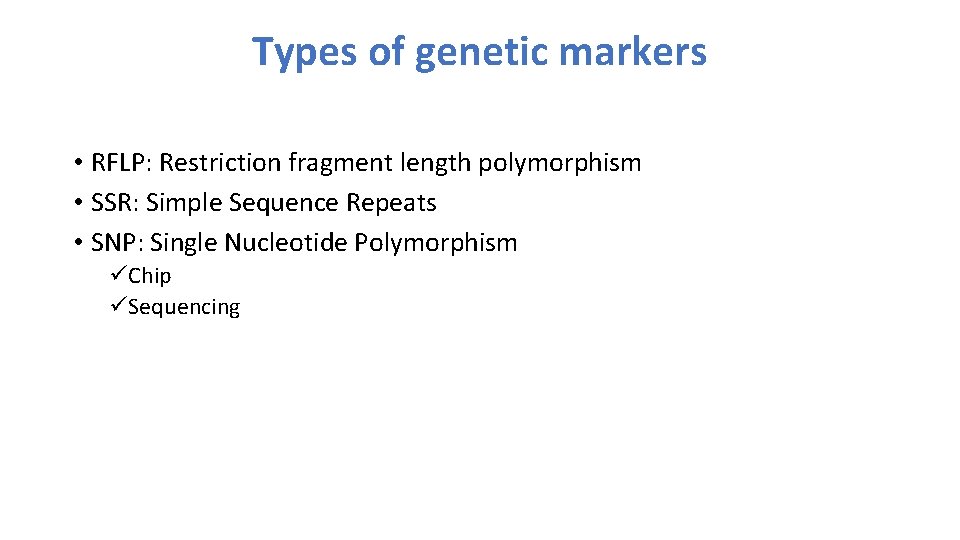
Types of genetic markers • RFLP: Restriction fragment length polymorphism • SSR: Simple Sequence Repeats • SNP: Single Nucleotide Polymorphism üChip üSequencing
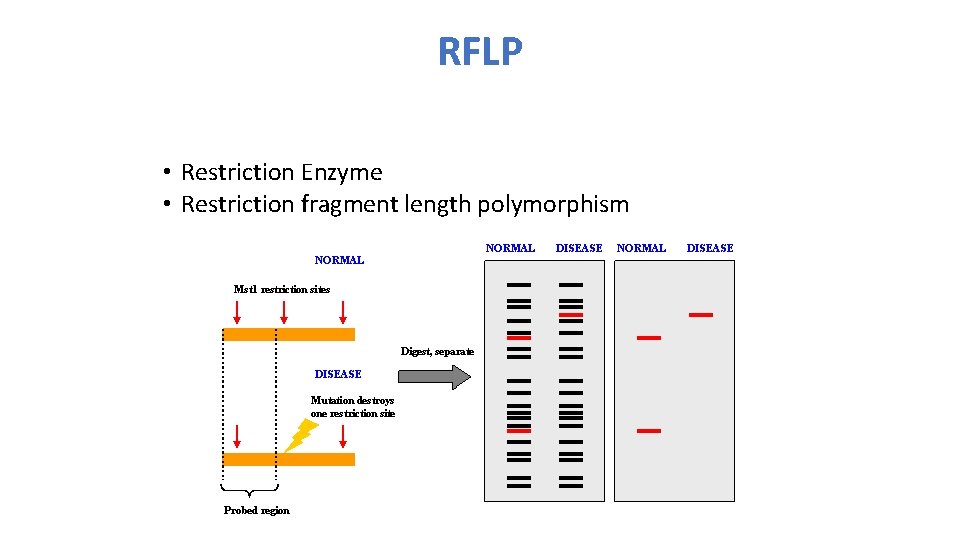
RFLP • Restriction Enzyme • Restriction fragment length polymorphism
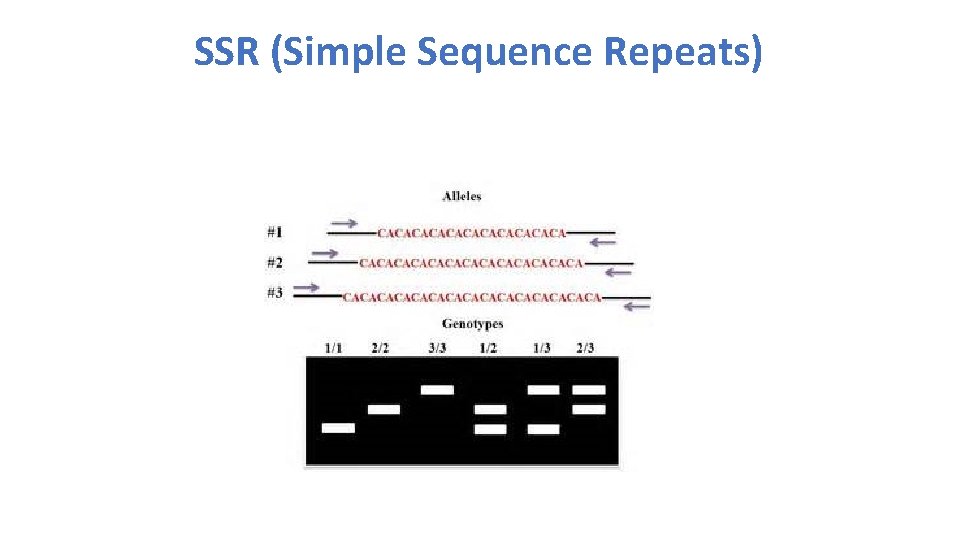
SSR (Simple Sequence Repeats)
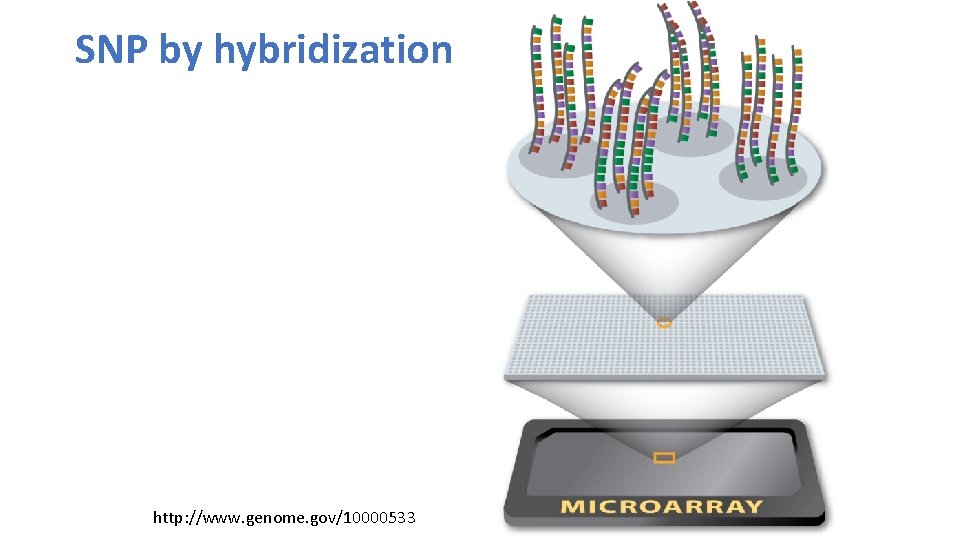
SNP by hybridization http: //www. genome. gov/10000533
Fredric Sanger • 1958 Nobel Price of Chemistry for Protein identification by electrophoresis • 1980 Nobel Price of Chemistry for DNA sequencing
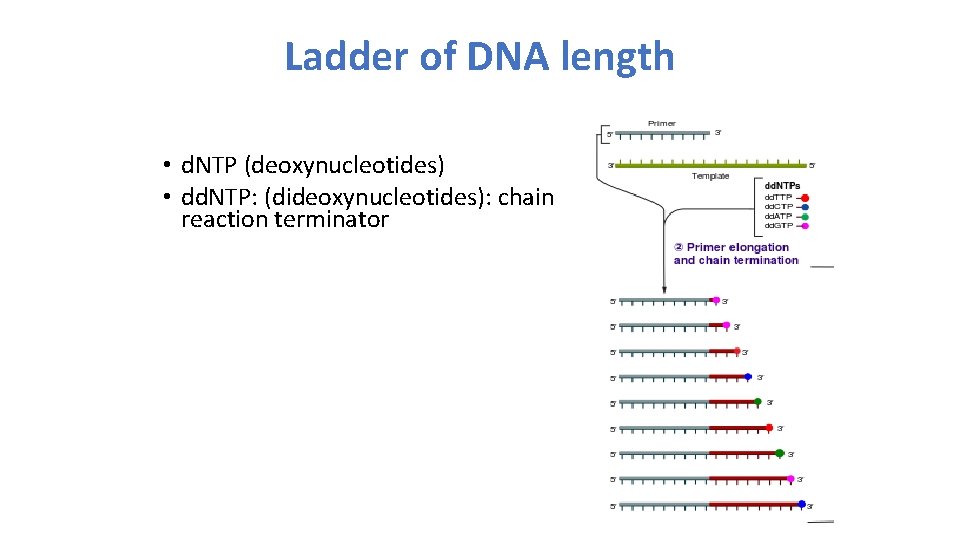
Ladder of DNA length • d. NTP (deoxynucleotides) • dd. NTP: (dideoxynucleotides): chain reaction terminator
1 st Generation DNA sequencing Fred Sanger and Alan R. Coulson, Nature 24, 687– 695 (1977)
2 nd generation sequencing • Sequencing-by-synthesis by 454 Life Science: Margulies, M. et al. Nature 437, 376– 380 (2005). • Multiplex Polony sequencing by George M. Church lab at Harvard Medical School: Shendure, J. et al. Science 309, 1728– 1732 (2005). 1 2 3 4 5 6
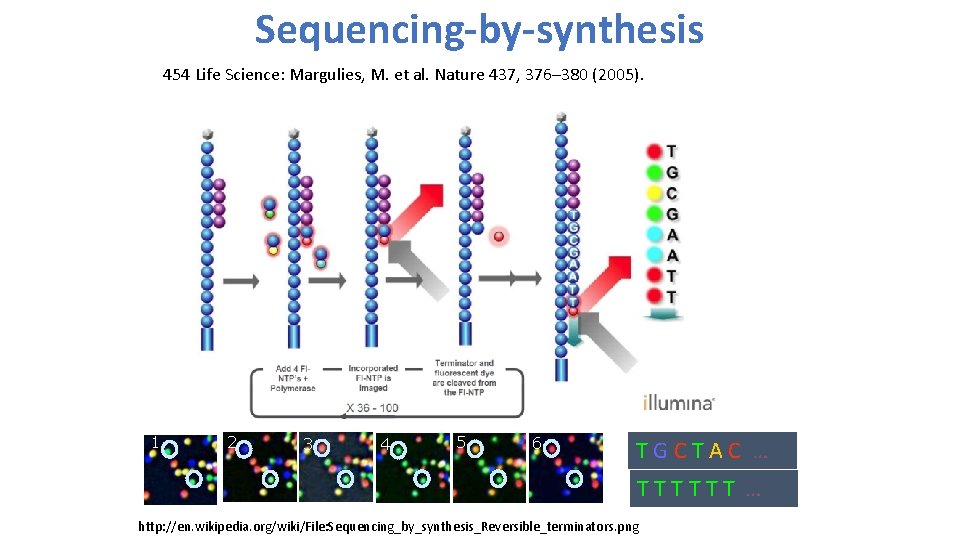
Sequencing-by-synthesis 454 Life Science: Margulies, M. et al. Nature 437, 376– 380 (2005). 1 2 3 4 5 6 TGCTAC … TTTTTT … http: //en. wikipedia. org/wiki/File: Sequencing_by_synthesis_Reversible_terminators. png

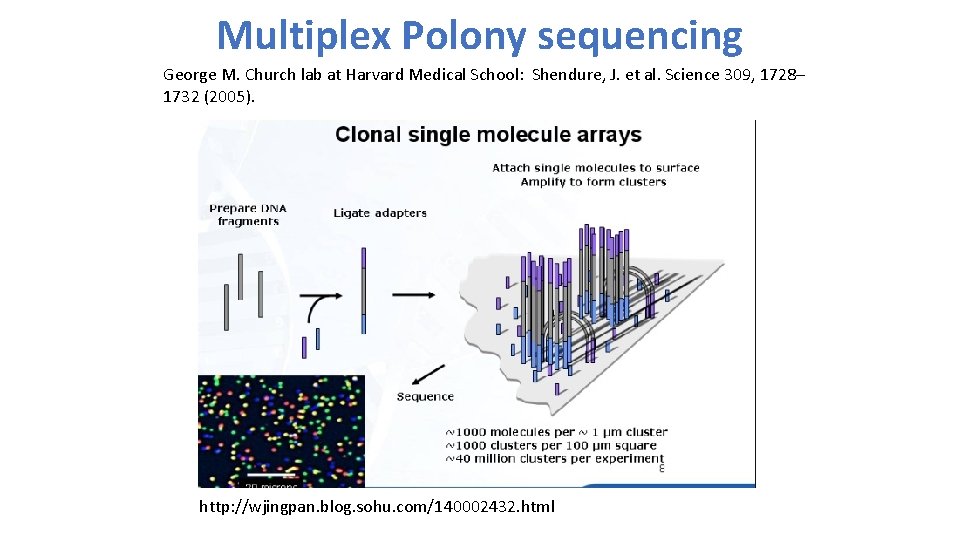
Multiplex Polony sequencing George M. Church lab at Harvard Medical School: Shendure, J. et al. Science 309, 1728– 1732 (2005). http: //wjingpan. blog. sohu. com/140002432. html
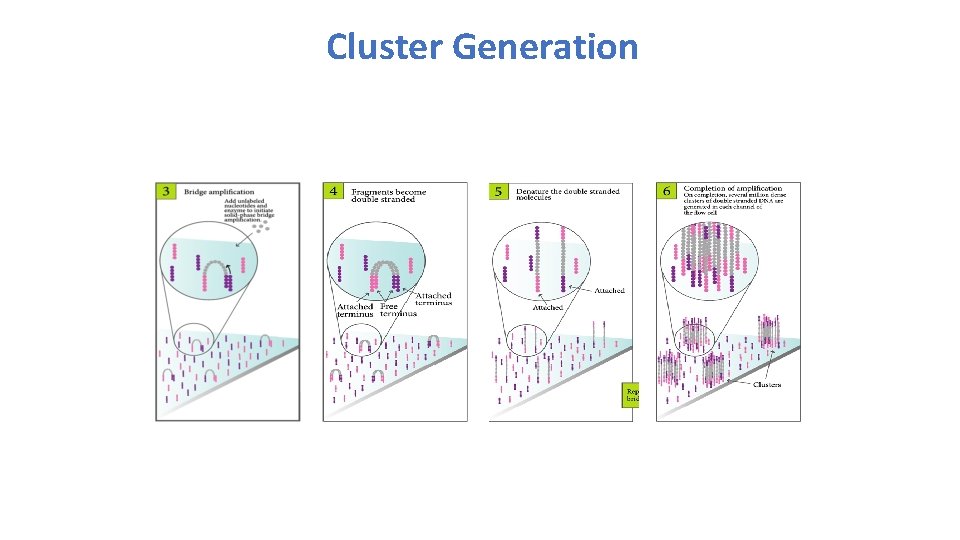
Cluster Generation
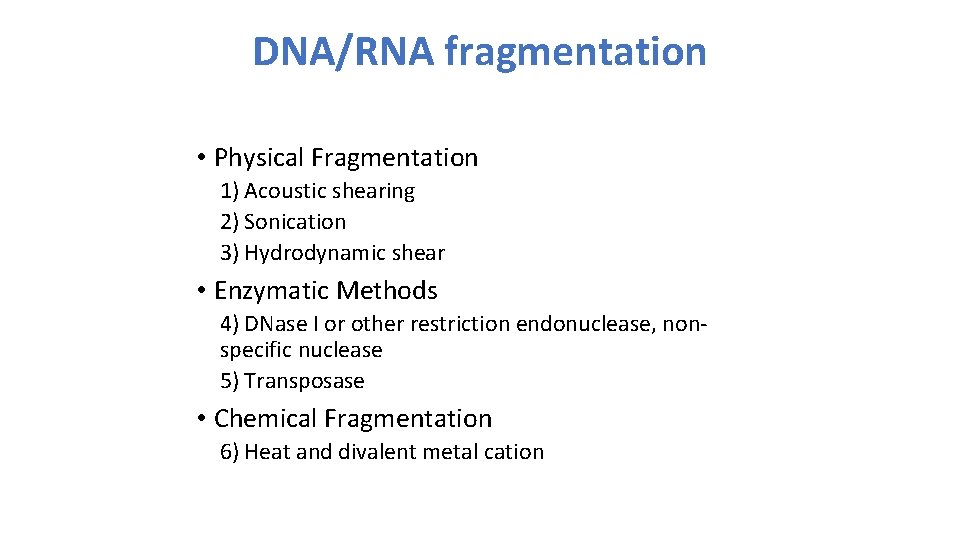
DNA/RNA fragmentation • Physical Fragmentation 1) Acoustic shearing 2) Sonication 3) Hydrodynamic shear • Enzymatic Methods 4) DNase I or other restriction endonuclease, nonspecific nuclease 5) Transposase • Chemical Fragmentation 6) Heat and divalent metal cation
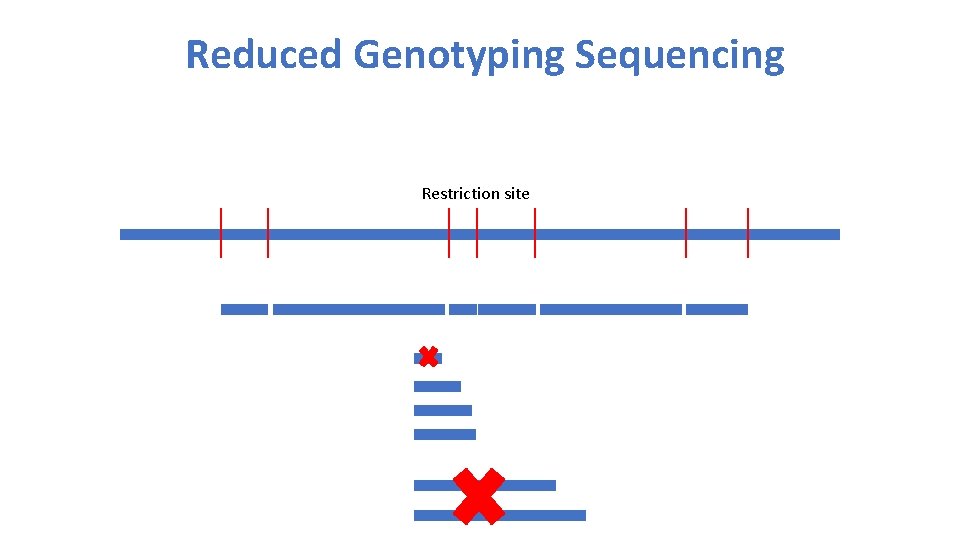
Reduced Genotyping Sequencing Restriction site
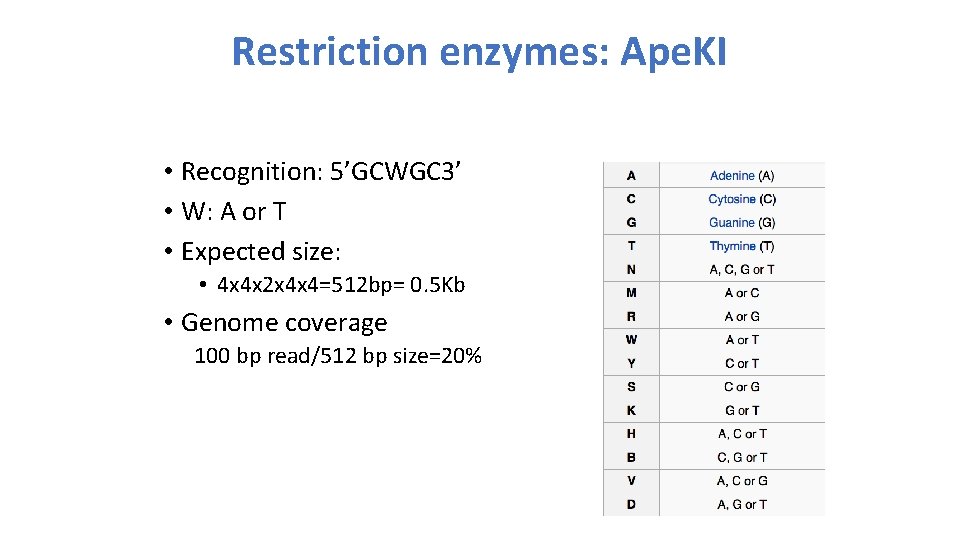
Restriction enzymes: Ape. KI • Recognition: 5’GCWGC 3’ • W: A or T • Expected size: • 4 x 4 x 2 x 4 x 4=512 bp= 0. 5 Kb • Genome coverage 100 bp read/512 bp size=20%

Restriction enzymes: Pst. I • Recognition: 5’ CTGCAG 3’ • Expected size: 4^6=4096 bp= 4 Kb • Genome coverage 100 bp read/4096 bp size=2. 5%
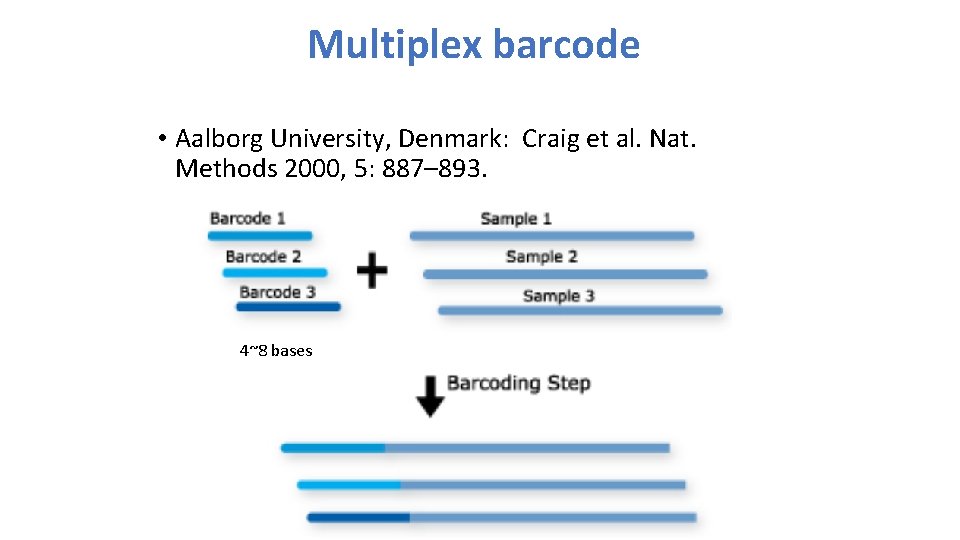
Multiplex barcode • Aalborg University, Denmark: Craig et al. Nat. Methods 2000, 5: 887– 893. 4~8 bases
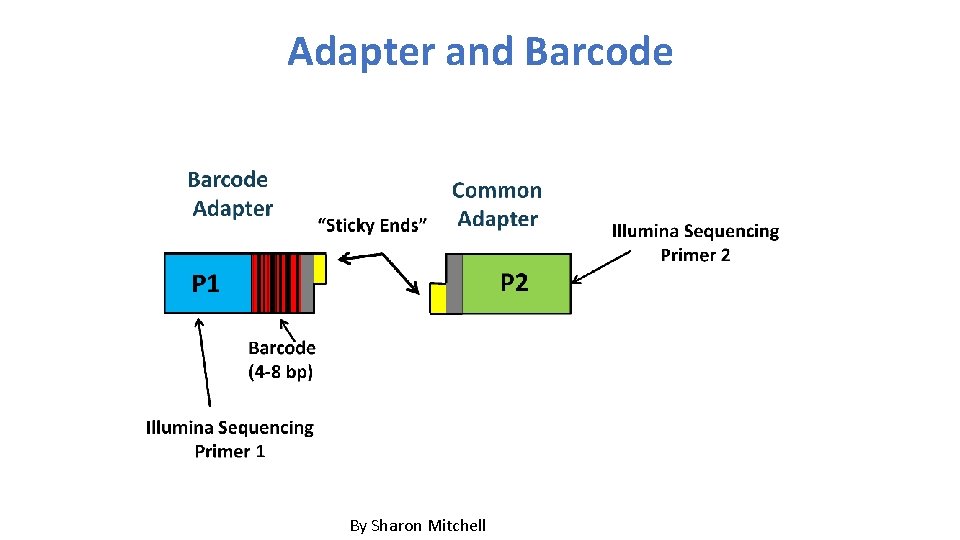
Adapter and Barcode By Sharon Mitchell
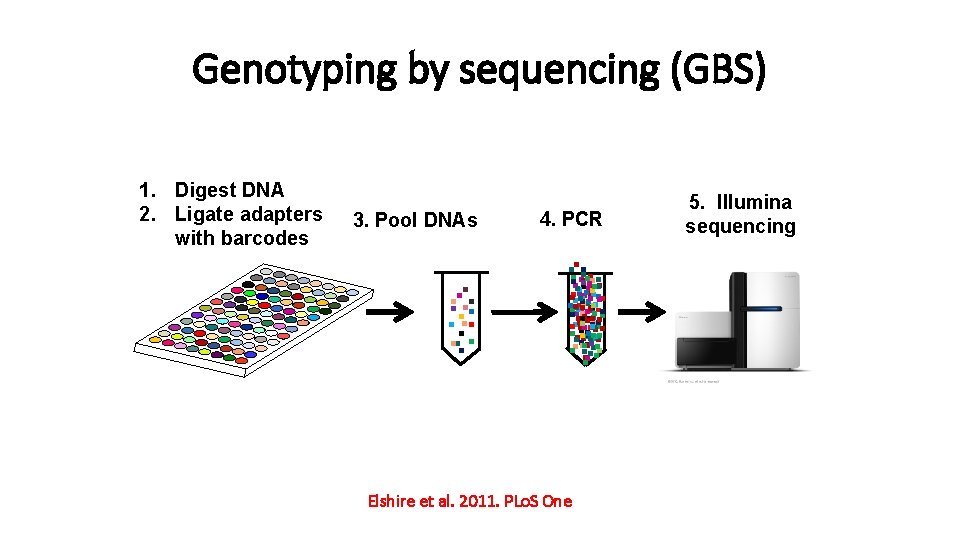
Genotyping by sequencing (GBS) 3. Pool DNAs 4. PCR . . . . . 1. Digest DNA 2. Ligate adapters with barcodes Elshire et al. 2011. PLo. S One 5. Illumina sequencing
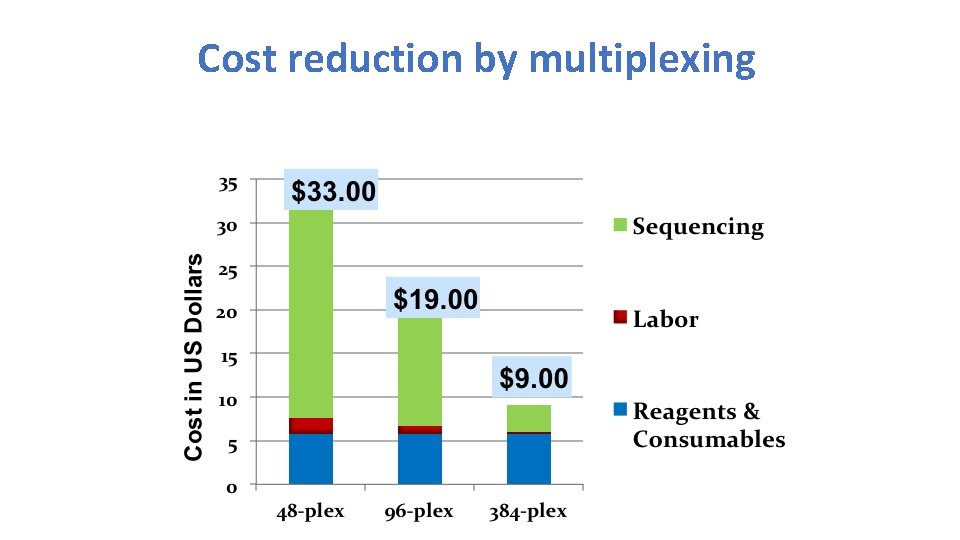
Cost reduction by multiplexing

Sequencing depth • Definition: Expected sequencing times per base pair • Calculation Ø 100 Mb genome, physically fragmented, 100 M read of 100 bp: 100 X Ø 3 G genome, 1% reduced by restriction enzyme, 50 multiplex, 6 G data (1 byte one base): 6 G/(50 x 3 Gx 1%)=4 X
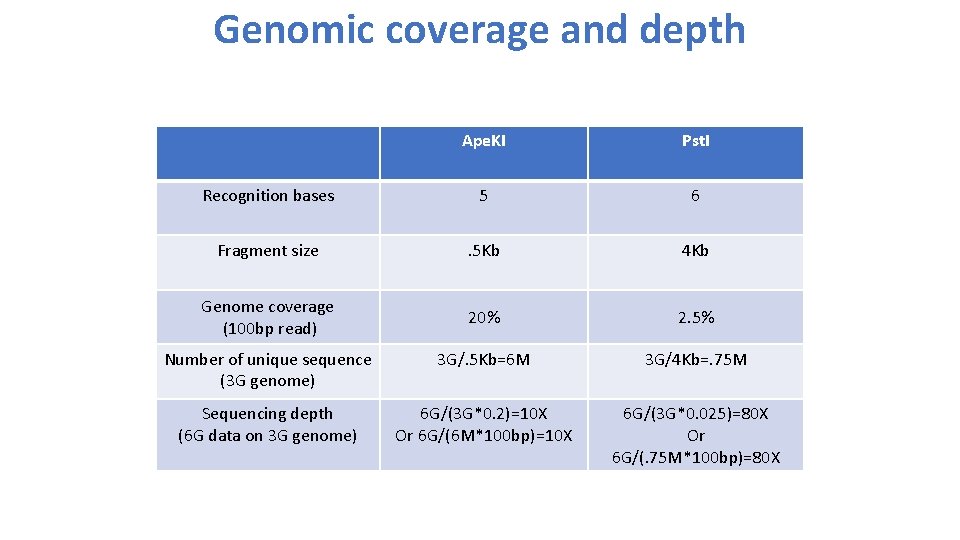
Genomic coverage and depth Ape. KI Pst. I Recognition bases 5 6 Fragment size . 5 Kb 4 Kb Genome coverage (100 bp read) 20% 2. 5% Number of unique sequence (3 G genome) 3 G/. 5 Kb=6 M 3 G/4 Kb=. 75 M Sequencing depth (6 G data on 3 G genome) 6 G/(3 G*0. 2)=10 X Or 6 G/(6 M*100 bp)=10 X 6 G/(3 G*0. 025)=80 X Or 6 G/(. 75 M*100 bp)=80 X
Distribution of length
![Distribution of length n=100000 #bumber of cuts size=30000 x=runif(n, 1, size) y=sort(x) interval=y[-1]-y[-n] hist(interval) Distribution of length n=100000 #bumber of cuts size=30000 x=runif(n, 1, size) y=sort(x) interval=y[-1]-y[-n] hist(interval)](http://slidetodoc.com/presentation_image_h2/6679955d7e4b8498babb825e496d4354/image-29.jpg)
Distribution of length n=100000 #bumber of cuts size=30000 x=runif(n, 1, size) y=sort(x) interval=y[-1]-y[-n] hist(interval) Ex=size/n Va=Ex*Ex m=mean(interval) v=var(interval) m v • Expectation of length=length/number of cut • Variance=Squared Expectation (need proof)
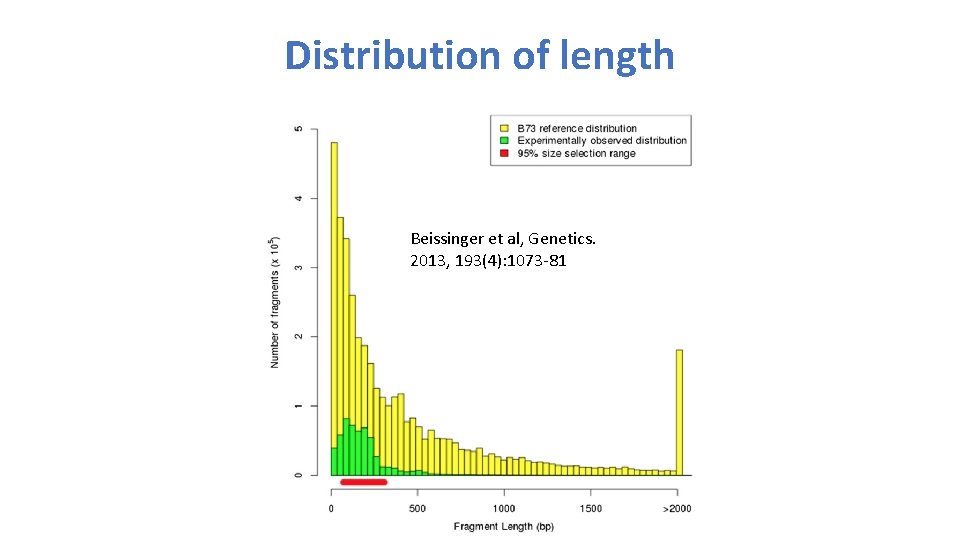
Distribution of length Beissinger et al, Genetics. 2013, 193(4): 1073 -81
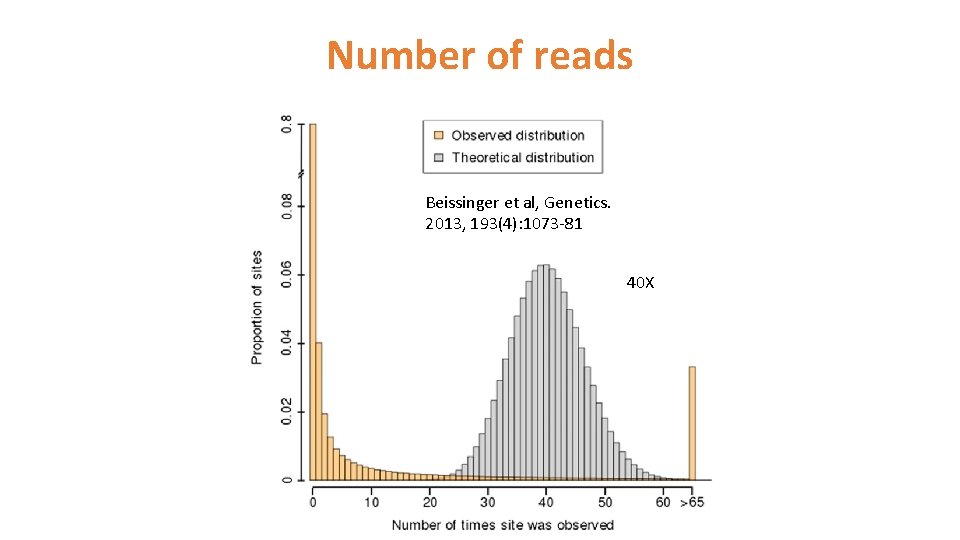
Number of reads Beissinger et al, Genetics. 2013, 193(4): 1073 -81 40 X
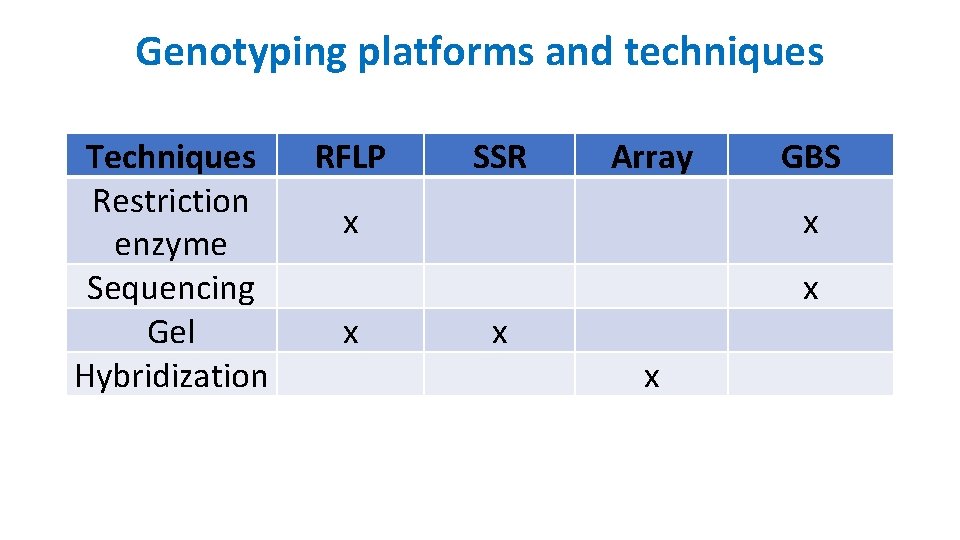
Genotyping platforms and techniques Techniques Restriction enzyme Sequencing Gel Hybridization RFLP SSR Array x GBS x x x

Outline • Genetic markers • Sequencing • Full vs. reduced
- Slides: 33