Statistical Genomics Lecture 11 Power type I error

![Bins (e. g. 100 Kb) big. Num=1 e 9 resolution=100000 bin=round((my. GM[, 2]*big. Num+my. Bins (e. g. 100 Kb) big. Num=1 e 9 resolution=100000 bin=round((my. GM[, 2]*big. Num+my.](https://slidetodoc.com/presentation_image/3207a06ab2c7c3e2a5c6ede6d1a67717/image-5.jpg)
![Bins of QTNs QTN. bin=result[my. Sim$QTN. position, ] QTN. bin Bins of QTNs QTN. bin=result[my. Sim$QTN. position, ] QTN. bin](https://slidetodoc.com/presentation_image/3207a06ab2c7c3e2a5c6ede6d1a67717/image-6.jpg)
![Sorted bins of QTNs index. qtn. p=order(QTN. bin[, 4]) QTN. bin[index. qtn. p, ] Sorted bins of QTNs index. qtn. p=order(QTN. bin[, 4]) QTN. bin[index. qtn. p, ]](https://slidetodoc.com/presentation_image/3207a06ab2c7c3e2a5c6ede6d1a67717/image-7.jpg)

![Replicates nrep=100 set. seed(99164) stat. Rep=replicate(nrep, { my. Sim=G 2 P(X=my. GD[, -1], h Replicates nrep=100 set. seed(99164) stat. Rep=replicate(nrep, { my. Sim=G 2 P(X=my. GD[, -1], h](https://slidetodoc.com/presentation_image/3207a06ab2c7c3e2a5c6ede6d1a67717/image-13.jpg)
![Means over replicates power=stat. Rep[[2]] #FDR s. fdr=seq(3, length(stat. Rep), 7) fdr=stat. Rep[s. fdr] Means over replicates power=stat. Rep[[2]] #FDR s. fdr=seq(3, length(stat. Rep), 7) fdr=stat. Rep[s. fdr]](https://slidetodoc.com/presentation_image/3207a06ab2c7c3e2a5c6ede6d1a67717/image-15.jpg)
![Plots of power vs. FDR the. Color=rainbow(4) plot(fdr. mean[, 1], power , type="b", col=the. Plots of power vs. FDR the. Color=rainbow(4) plot(fdr. mean[, 1], power , type="b", col=the.](https://slidetodoc.com/presentation_image/3207a06ab2c7c3e2a5c6ede6d1a67717/image-16.jpg)

![Means and plot power 25=stat. Rep 25[[2]] s. t 1=seq(4, length(stat. Rep 25), 7) Means and plot power 25=stat. Rep 25[[2]] s. t 1=seq(4, length(stat. Rep 25), 7)](https://slidetodoc.com/presentation_image/3207a06ab2c7c3e2a5c6ede6d1a67717/image-20.jpg)
- Slides: 21

Statistical Genomics Lecture 11: Power, type I error and FDR Zhiwu Zhang Washington State University

Outline • • Simulation of phenotype from genotype GWAS by correlation Power FDR Cutoff Null distribution of p values Resolution QTN bins and non-QTN bins

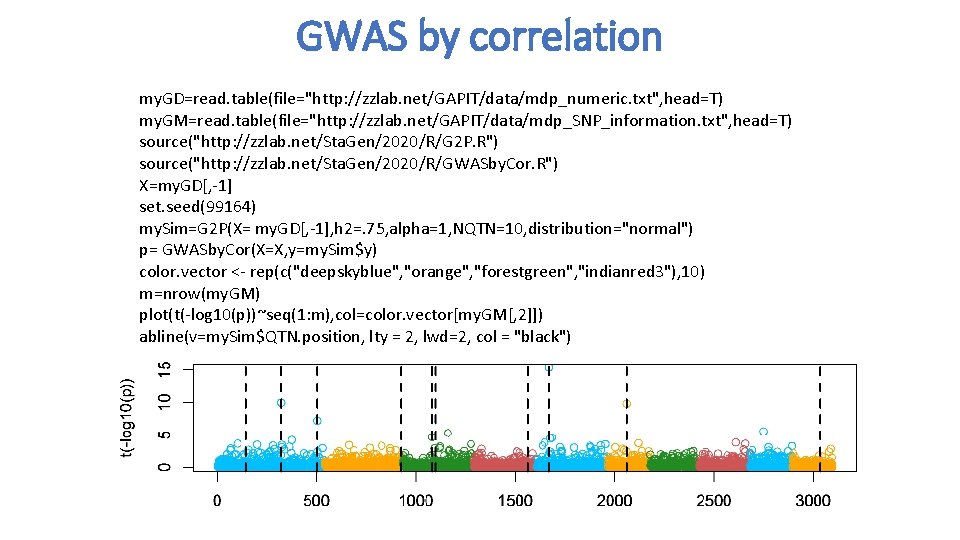
GWAS by correlation my. GD=read. table(file="http: //zzlab. net/GAPIT/data/mdp_numeric. txt", head=T) my. GM=read. table(file="http: //zzlab. net/GAPIT/data/mdp_SNP_information. txt", head=T) source("http: //zzlab. net/Sta. Gen/2020/R/G 2 P. R") source("http: //zzlab. net/Sta. Gen/2020/R/GWASby. Cor. R") X=my. GD[, -1] set. seed(99164) my. Sim=G 2 P(X= my. GD[, -1], h 2=. 75, alpha=1, NQTN=10, distribution="normal") p= GWASby. Cor(X=X, y=my. Sim$y) color. vector <- rep(c("deepskyblue", "orange", "forestgreen", "indianred 3"), 10) m=nrow(my. GM) plot(t(-log 10(p))~seq(1: m), col=color. vector[my. GM[, 2]]) abline(v=my. Sim$QTN. position, lty = 2, lwd=2, col = "black")

Resolution and bin approach • 10 Kb is really good, 100 Kb is OK • Bins with QTNs for power • Bins without QTNs for type I error
![Bins e g 100 Kb big Num1 e 9 resolution100000 binroundmy GM 2big Nummy Bins (e. g. 100 Kb) big. Num=1 e 9 resolution=100000 bin=round((my. GM[, 2]*big. Num+my.](https://slidetodoc.com/presentation_image/3207a06ab2c7c3e2a5c6ede6d1a67717/image-5.jpg)
Bins (e. g. 100 Kb) big. Num=1 e 9 resolution=100000 bin=round((my. GM[, 2]*big. Num+my. GM[, 3])/resolution) result=cbind(my. GM, t(p), bin) head(result) Minimum p value within bin
![Bins of QTNs QTN binresultmy SimQTN position QTN bin Bins of QTNs QTN. bin=result[my. Sim$QTN. position, ] QTN. bin](https://slidetodoc.com/presentation_image/3207a06ab2c7c3e2a5c6ede6d1a67717/image-6.jpg)
Bins of QTNs QTN. bin=result[my. Sim$QTN. position, ] QTN. bin
![Sorted bins of QTNs index qtn porderQTN bin 4 QTN binindex qtn p Sorted bins of QTNs index. qtn. p=order(QTN. bin[, 4]) QTN. bin[index. qtn. p, ]](https://slidetodoc.com/presentation_image/3207a06ab2c7c3e2a5c6ede6d1a67717/image-7.jpg)
Sorted bins of QTNs index. qtn. p=order(QTN. bin[, 4]) QTN. bin[index. qtn. p, ]

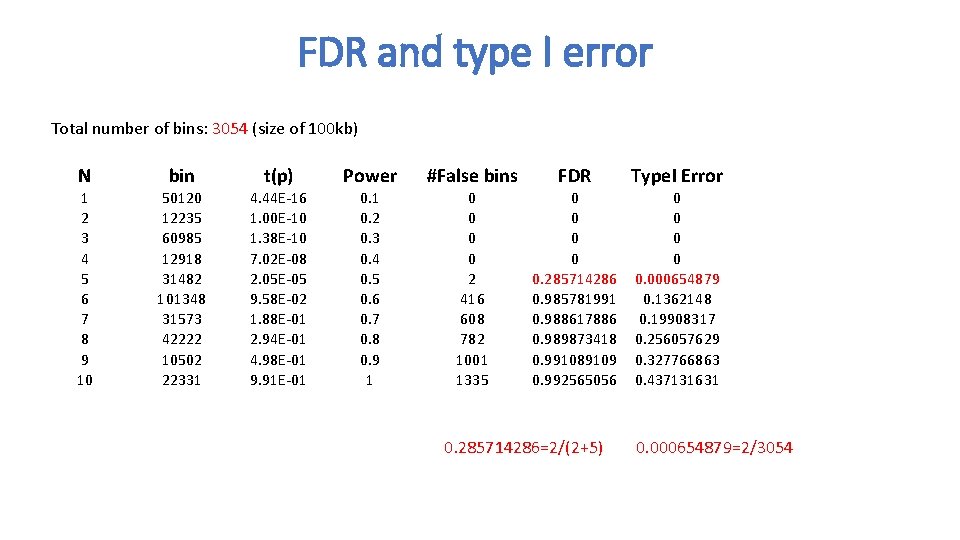
FDR and type I error Total number of bins: 3054 (size of 100 kb) N bin t(p) Power #False bins FDR Type. I Error 1 2 3 4 5 6 7 8 9 10 50120 12235 60985 12918 31482 101348 31573 42222 10502 22331 4. 44 E-16 1. 00 E-10 1. 38 E-10 7. 02 E-08 2. 05 E-05 9. 58 E-02 1. 88 E-01 2. 94 E-01 4. 98 E-01 9. 91 E-01 0. 2 0. 3 0. 4 0. 5 0. 6 0. 7 0. 8 0. 9 1 0 0 2 416 608 782 1001 1335 0 0 0. 285714286 0. 985781991 0. 988617886 0. 989873418 0. 991089109 0. 992565056 0 0 0. 000654879 0. 1362148 0. 19908317 0. 256057629 0. 327766863 0. 437131631 0. 285714286=2/(2+5) 0. 000654879=2/3054

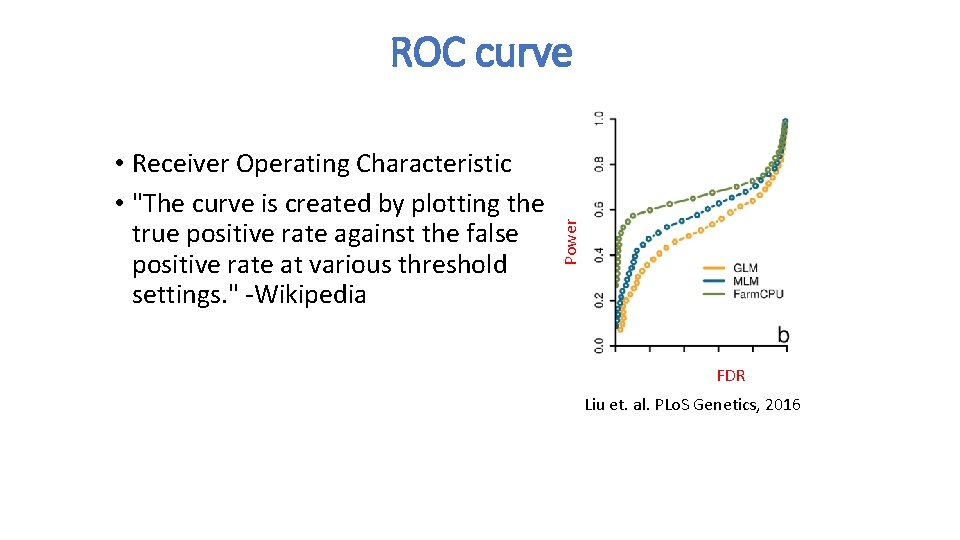
• Receiver Operating Characteristic • "The curve is created by plotting the true positive rate against the false positive rate at various threshold settings. " -Wikipedia Power ROC curve FDR Liu et. al. PLo. S Genetics, 2016

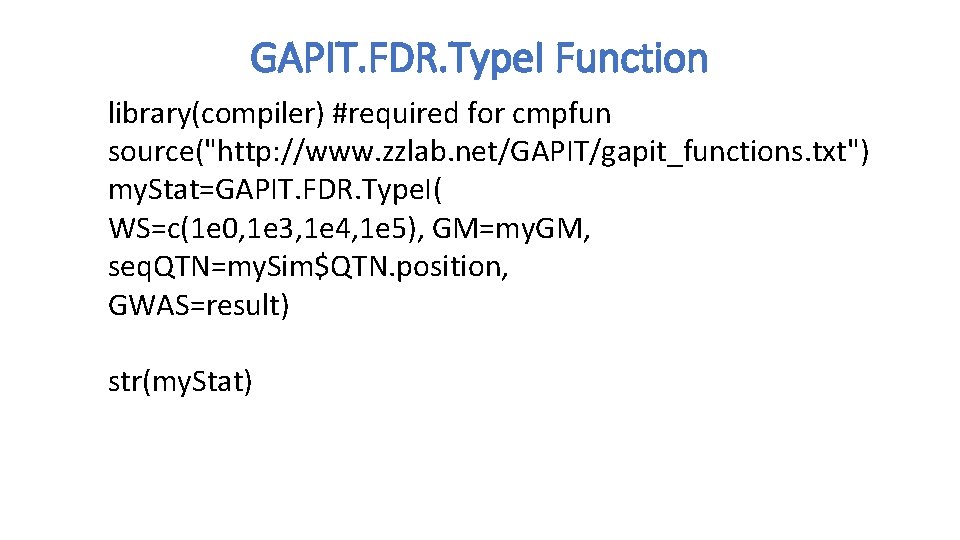
GAPIT. FDR. Type. I Function library(compiler) #required for cmpfun source("http: //www. zzlab. net/GAPIT/gapit_functions. txt") my. Stat=GAPIT. FDR. Type. I( WS=c(1 e 0, 1 e 3, 1 e 4, 1 e 5), GM=my. GM, seq. QTN=my. Sim$QTN. position, GWAS=result) str(my. Stat)

Return

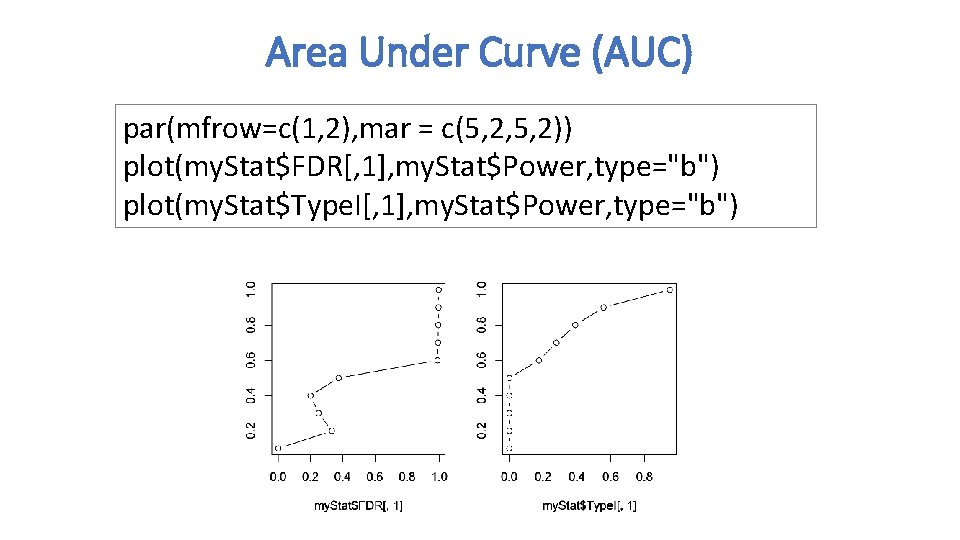
Area Under Curve (AUC) par(mfrow=c(1, 2), mar = c(5, 2, 5, 2)) plot(my. Stat$FDR[, 1], my. Stat$Power, type="b") plot(my. Stat$Type. I[, 1], my. Stat$Power, type="b")
![Replicates nrep100 set seed99164 stat Repreplicatenrep my SimG 2 PXmy GD 1 h Replicates nrep=100 set. seed(99164) stat. Rep=replicate(nrep, { my. Sim=G 2 P(X=my. GD[, -1], h](https://slidetodoc.com/presentation_image/3207a06ab2c7c3e2a5c6ede6d1a67717/image-13.jpg)
Replicates nrep=100 set. seed(99164) stat. Rep=replicate(nrep, { my. Sim=G 2 P(X=my. GD[, -1], h 2=. 5, alpha=1, NQTN=10, distribution="norm") p=p= GWASby. Cor(X=my. GD[, -1], y=my. Sim$y) seq. QTN=my. Sim$QTN. position my. GWAS=cbind(my. GM, t(p), NA) my. Stat=GAPIT. FDR. Type. I(WS=c(1 e 0, 1 e 3, 1 e 4, 1 e 5), GM=my. GM, seq. QTN=my. Sim$QTN. position, GWAS=my. GWAS, max. Out=100, Max. BP=1 e 10) })

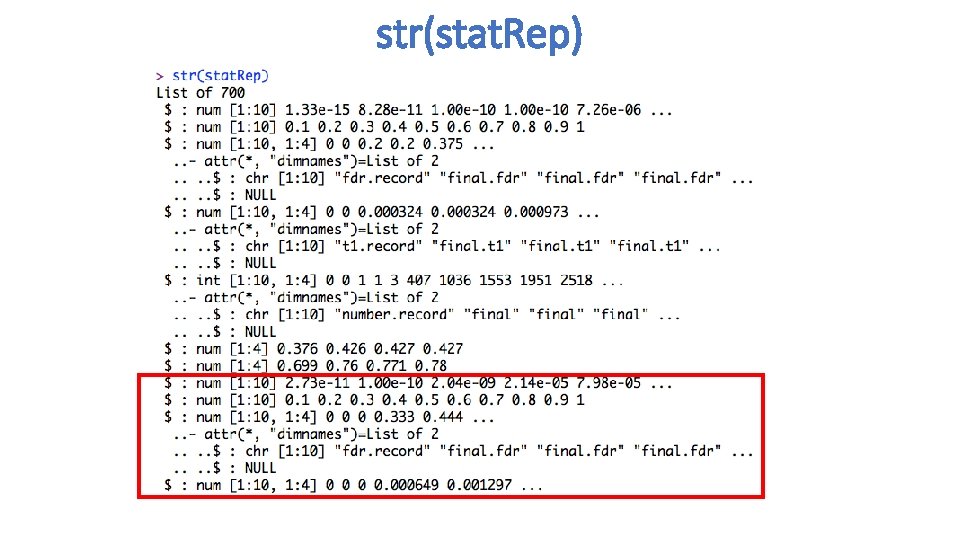
str(stat. Rep)
![Means over replicates powerstat Rep2 FDR s fdrseq3 lengthstat Rep 7 fdrstat Reps fdr Means over replicates power=stat. Rep[[2]] #FDR s. fdr=seq(3, length(stat. Rep), 7) fdr=stat. Rep[s. fdr]](https://slidetodoc.com/presentation_image/3207a06ab2c7c3e2a5c6ede6d1a67717/image-15.jpg)
Means over replicates power=stat. Rep[[2]] #FDR s. fdr=seq(3, length(stat. Rep), 7) fdr=stat. Rep[s. fdr] fdr. mean=Reduce ("+", fdr) / length(fdr) #AUC: power vs. FDR s. auc. fdr=seq(6, length(stat. Rep), 7) auc. fdr=stat. Rep[s. auc. fdr] auc. fdr. mean=Reduce ("+", auc. fdr) / length(auc. fdr)
![Plots of power vs FDR the Colorrainbow4 plotfdr mean 1 power typeb colthe Plots of power vs. FDR the. Color=rainbow(4) plot(fdr. mean[, 1], power , type="b", col=the.](https://slidetodoc.com/presentation_image/3207a06ab2c7c3e2a5c6ede6d1a67717/image-16.jpg)
Plots of power vs. FDR the. Color=rainbow(4) plot(fdr. mean[, 1], power , type="b", col=the. Color [1], xlim=c(0, 1)) for(i in 2: ncol(fdr. mean)){ lines(fdr. mean[, i], power , type="b", col= the. Color [i]) }

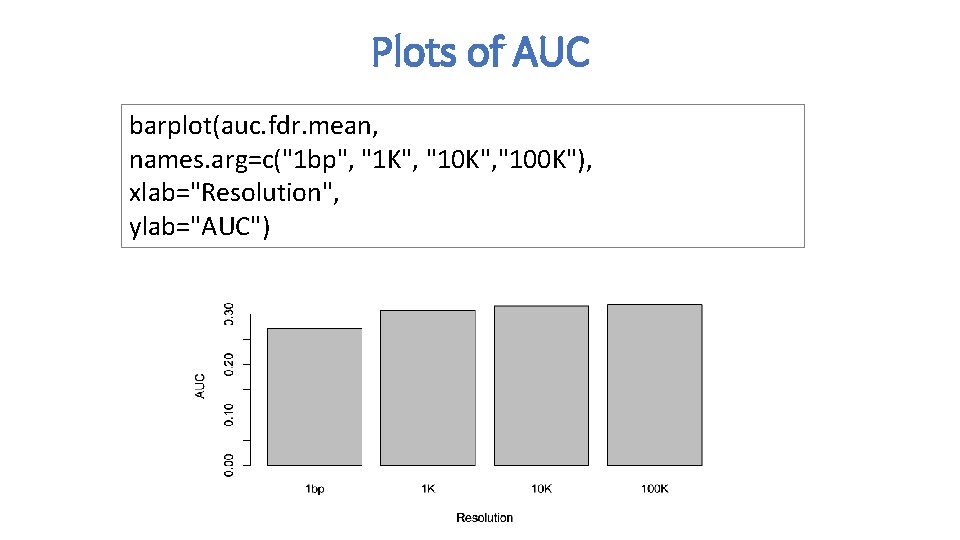
Plots of AUC barplot(auc. fdr. mean, names. arg=c("1 bp", "1 K", "100 K"), xlab="Resolution", ylab="AUC")

ROC with different heritability • h 2= 25% vs. 75% • 10 QTNs • Normal distributed QTN effect • 100 kb resolution • Power against Type I error

Simulation and GWAS nrep=100 set. seed(99164) #h 2=25% stat. Rep 25=replicate(nrep, { my. Sim=G 2 P(X=my. GD[, -1], h 2=. 25, alpha=1, NQTN=10, distribution="norm") p=p= GWASby. Cor(X=my. GD[, -1], y=my. Sim$y) seq. QTN=my. Sim$QTN. position my. GWAS=cbind(my. GM, t(p), NA) my. Stat=GAPIT. FDR. Type. I(WS=c(1 e 0, 1 e 3, 1 e 4, 1 e 5), GM=my. GM, seq. QTN=my. Sim$QTN. position, GWAS=my. GWAS, max. Out=100, Max. BP=1 e 10)}) #h 2=75% stat. Rep 75=replicate(nrep, { my. Sim=G 2 P(X=my. GD[, -1], h 2=. 75, alpha=1, NQTN=10, distribution="norm") p= GWASby. Cor(X=my. GD[, -1], y=my. Sim$y) seq. QTN=my. Sim$QTN. position my. GWAS=cbind(my. GM, t(p), NA) my. Stat=GAPIT. FDR. Type. I(WS=c(1 e 0, 1 e 3, 1 e 4, 1 e 5), GM=my. GM, seq. QTN=my. Sim$QTN. position, GWAS=my. GWAS, max. Out=100, Max. BP=1 e 10)})
![Means and plot power 25stat Rep 252 s t 1seq4 lengthstat Rep 25 7 Means and plot power 25=stat. Rep 25[[2]] s. t 1=seq(4, length(stat. Rep 25), 7)](https://slidetodoc.com/presentation_image/3207a06ab2c7c3e2a5c6ede6d1a67717/image-20.jpg)
Means and plot power 25=stat. Rep 25[[2]] s. t 1=seq(4, length(stat. Rep 25), 7) t 1=stat. Rep 25[s. t 1] t 1. mean. 25=Reduce ("+", t 1) / length(t 1) power 75=stat. Rep 75[[2]] s. t 1=seq(4, length(stat. Rep 75), 7) t 1=stat. Rep 75[s. t 1] t 1. mean. 75=Reduce ("+", t 1) / length(t 1) plot(t 1. mean. 25[, 4], power 25, type="b", col="blue", xlim=c(0, 0. 6)) lines(t 1. mean. 75[, 4], power 75, type="b", col= "red")

Outline • • Simulation of phenotype from genotype GWAS by correlation Power FDR Cutoff Null distribution of p values Resolution QTN bins and non-QTN bins