State University of Campinas UNICAMP School of Medicine








- Slides: 28

State University of Campinas (UNICAMP) School of Medicine Hematology and Transfusion Medicine Center - Hemocentro Screening of Genetic Variants in Familial Case of Myeloid Neoplasm Using Exome Sequencing Sara Teresinha Olalla Saad Campinas – São Paulo, Brazil 2016


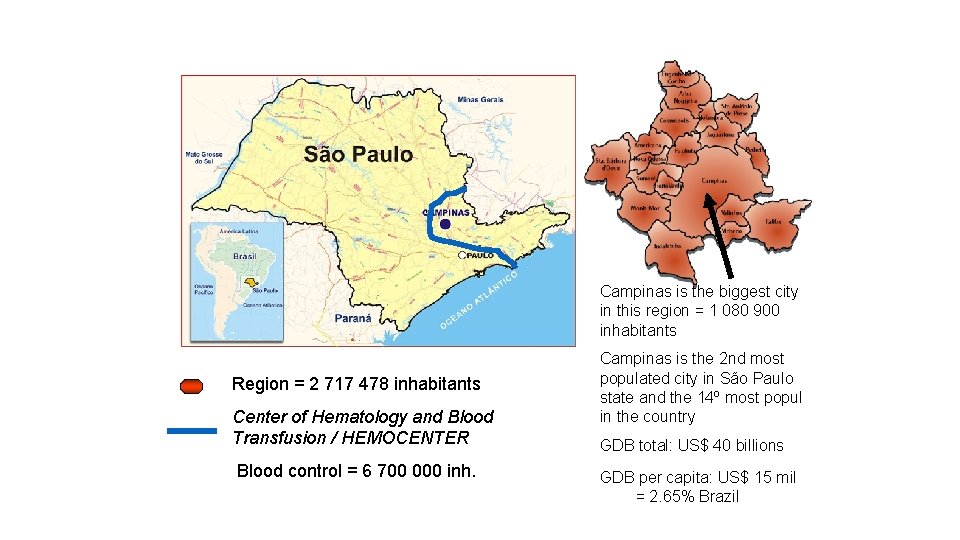
Campinas is the biggest city in this region = 1 080 900 inhabitants Region = 2 717 478 inhabitants Center of Hematology and Blood Transfusion / HEMOCENTER Blood control = 6 700 000 inh. Campinas is the 2 nd most populated city in São Paulo state and the 14º most popul in the country GDB total: US$ 40 billions GDB per capita: US$ 15 mil = 2. 65% Brazil

Bone marrow

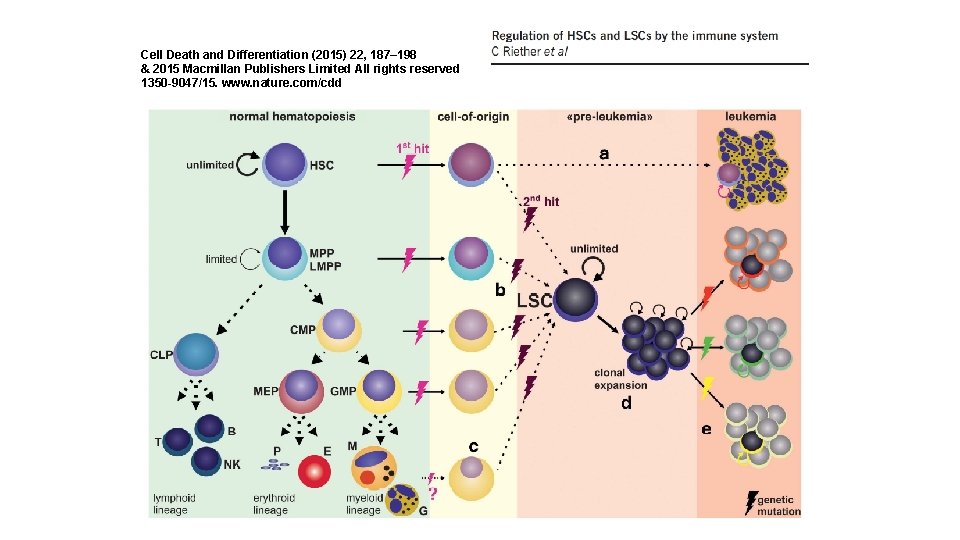
Cell Death and Differentiation (2015) 22, 187– 198 & 2015 Macmillan Publishers Limited All rights reserved 1350 -9047/15. www. nature. com/cdd

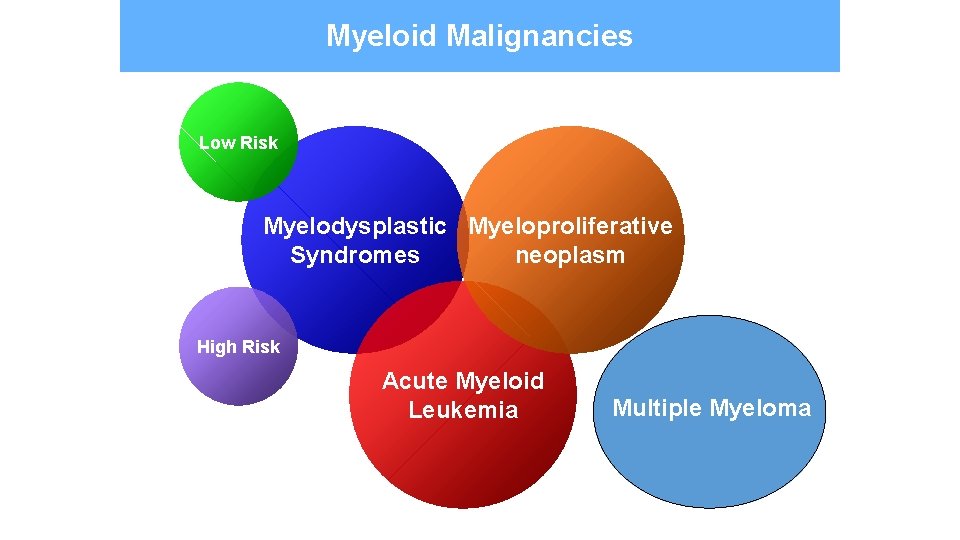
Myeloid Malignancies Low Risk Myelodysplastic Myeloproliferative Syndromes neoplasm High Risk Acute Myeloid Leukemia Multiple Myeloma

AIM • General To investigate and to evaluate possible somatic and germline mutations in a specific group of patients with familial and atypical myeloid neoplasms.

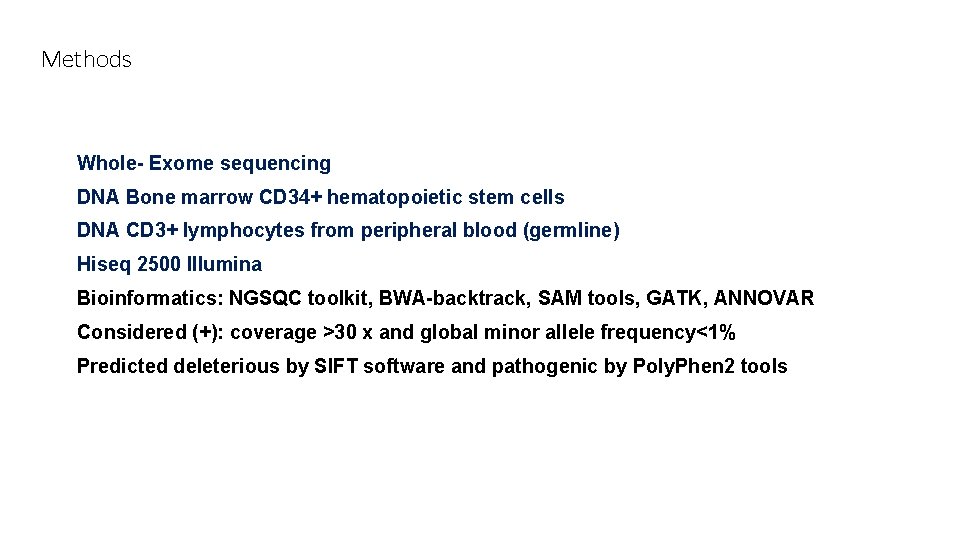
Methods Whole- Exome sequencing DNA Bone marrow CD 34+ hematopoietic stem cells DNA CD 3+ lymphocytes from peripheral blood (germline) Hiseq 2500 Illumina Bioinformatics: NGSQC toolkit, BWA-backtrack, SAM tools, GATK, ANNOVAR Considered (+): coverage >30 x and global minor allele frequency<1% Predicted deleterious by SIFT software and pathogenic by Poly. Phen 2 tools

Case 1 I-1 III-1 III-2 III-3 IV-1 2 IV- I-2 III-4 IV-3 IV-4 V-1 III-5 IV-5 III-6 Two sisters, 39 and 40, with primary myelofibrosis; History of longtime exposure to pesticides DDT-type; BM: hypercellularity and myelofibrosis; PB: leukocytosis, neutrophilia and thrombocytopenia; Symptomatic anemia; No presence of BCR-ABL 1 and JAK 2 V 617 F; The mother (II-2) and the youngest brother (III-6) → asymptomatic; Ø Maternal grandmother (I-2) → died due to uterine cancer; Ø Awaiting bone marrow transplantation. Ø Ø Ø Ø IV-6 Ø Oldest sister (III-3): § BM karyotype: 45, XX, -7 [35% metaphases]. Ø Youngest sister (III-4): § Normal karyotype (BM). Thrombocytopenia Uterine cancer

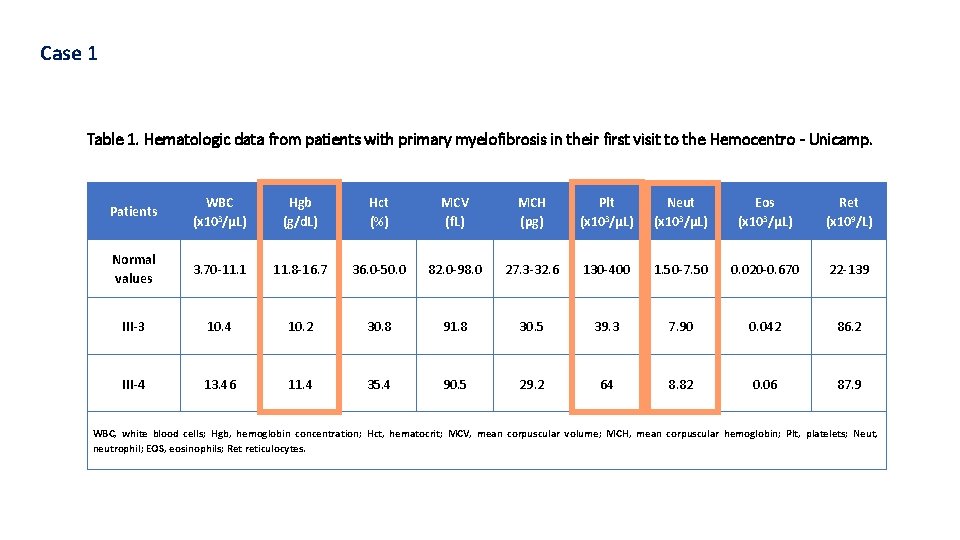
Case 1 Table 1. Hematologic data from patients with primary myelofibrosis in their first visit to the Hemocentro - Unicamp. Patients WBC (x 103/μL) Hgb (g/d. L) Hct (%) MCV (f. L) MCH (pg) Plt (x 103/μL) Neut (x 103/μL) Eos (x 103/μL) Ret (x 109/L) Normal values 3. 70 -11. 1 11. 8 -16. 7 36. 0 -50. 0 82. 0 -98. 0 27. 3 -32. 6 130 -400 1. 50 -7. 50 0. 020 -0. 670 22 -139 III-3 10. 4 10. 2 30. 8 91. 8 30. 5 39. 3 7. 90 0. 042 86. 2 III-4 13. 46 11. 4 35. 4 90. 5 29. 2 64 8. 82 0. 06 87. 9 WBC, white blood cells; Hgb, hemoglobin concentration; Hct, hematocrit; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; Plt, platelets; Neut, neutrophil; EOS, eosinophils; Ret reticulocytes.

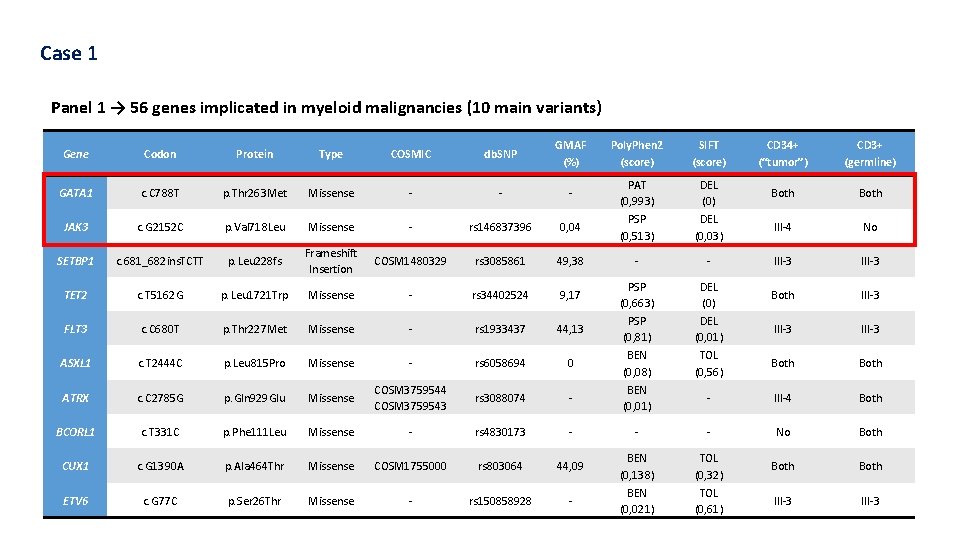
Case 1 Panel 1 → 56 genes implicated in myeloid malignancies (10 main variants) Gene Codon Protein Type COSMIC db. SNP GMAF (%) GATA 1 c. C 788 T p. Thr 263 Met Missense - - - JAK 3 c. G 2152 C p. Val 718 Leu Missense - rs 146837396 0, 04 SETBP 1 c. 681_682 ins. TCTT p. Leu 228 fs Frameshift Insertion COSM 1480329 rs 3085861 49, 38 TET 2 c. T 5162 G p. Leu 1721 Trp Missense - rs 34402524 9, 17 FLT 3 c. C 680 T p. Thr 227 Met Missense - rs 1933437 44, 13 ASXL 1 c. T 2444 C p. Leu 815 Pro Missense - rs 6058694 0 ATRX c. C 2785 G p. Gln 929 Glu Missense COSM 3759544 COSM 3759543 rs 3088074 - BCORL 1 c. T 331 C p. Phe 111 Leu Missense - rs 4830173 - CUX 1 c. G 1390 A p. Ala 464 Thr Missense COSM 1755000 rs 803064 44, 09 ETV 6 c. G 77 C p. Ser 26 Thr Missense - rs 150858928 - Poly. Phen 2 (score) SIFT (score) CD 34+ (“tumor”) CD 3+ (germline) PAT (0, 993) PSP (0, 513) DEL (0, 03) Both III-4 No - - III-3 PSP (0, 663) PSP (0, 81) BEN (0, 08) BEN (0, 01) DEL (0, 01) TOL (0, 56) Both III-3 Both - III-4 Both - - No Both BEN (0, 138) BEN (0, 021) TOL (0, 32) TOL (0, 61) Both III-3

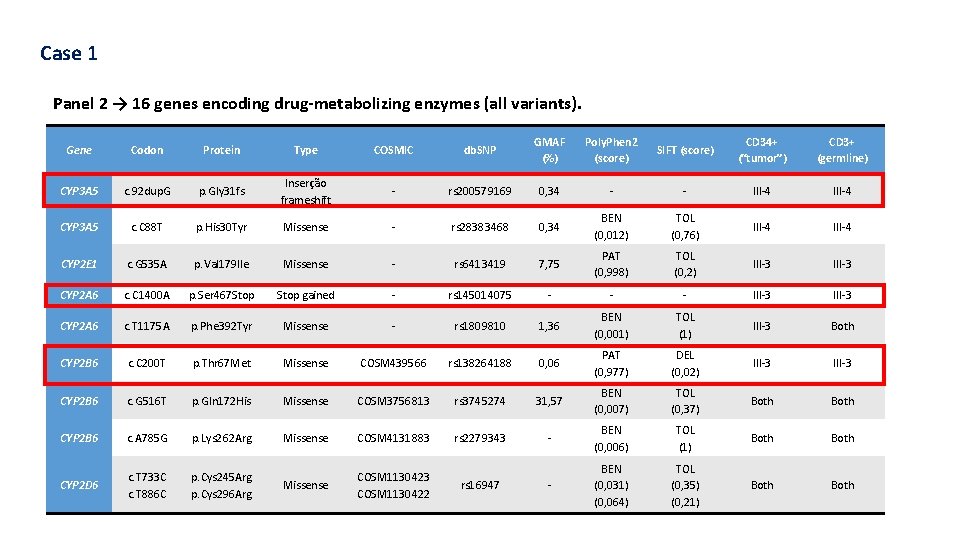
Case 1 Panel 2 → 16 genes encoding drug-metabolizing enzymes (all variants). Gene Codon Protein Type COSMIC db. SNP GMAF (%) Poly. Phen 2 (score) SIFT (score) CD 34+ (“tumor”) CD 3+ (germline) CYP 3 A 5 c. 92 dup. Gly 31 fs Inserção frameshift - rs 200579169 0, 34 - - III-4 CYP 3 A 5 c. C 88 T p. His 30 Tyr Missense - rs 28383468 0, 34 BEN (0, 012) TOL (0, 76) III-4 CYP 2 E 1 c. G 535 A p. Val 179 Ile Missense - rs 6413419 7, 75 PAT (0, 998) TOL (0, 2) III-3 CYP 2 A 6 c. C 1400 A p. Ser 467 Stop gained - rs 145014075 - - - III-3 CYP 2 A 6 c. T 1175 A p. Phe 392 Tyr Missense - rs 1809810 1, 36 BEN (0, 001) TOL (1) III-3 Both CYP 2 B 6 c. C 200 T p. Thr 67 Met Missense COSM 439566 rs 138264188 0, 06 PAT (0, 977) DEL (0, 02) III-3 CYP 2 B 6 c. G 516 T p. Gln 172 His Missense COSM 3756813 rs 3745274 31, 57 BEN (0, 007) TOL (0, 37) Both CYP 2 B 6 c. A 785 G p. Lys 262 Arg Missense COSM 4131883 rs 2279343 - BEN (0, 006) TOL (1) Both CYP 2 D 6 c. T 733 C c. T 886 C p. Cys 245 Arg p. Cys 296 Arg Missense COSM 1130423 COSM 1130422 - BEN (0, 031) (0, 064) TOL (0, 35) (0, 21) Both rs 16947

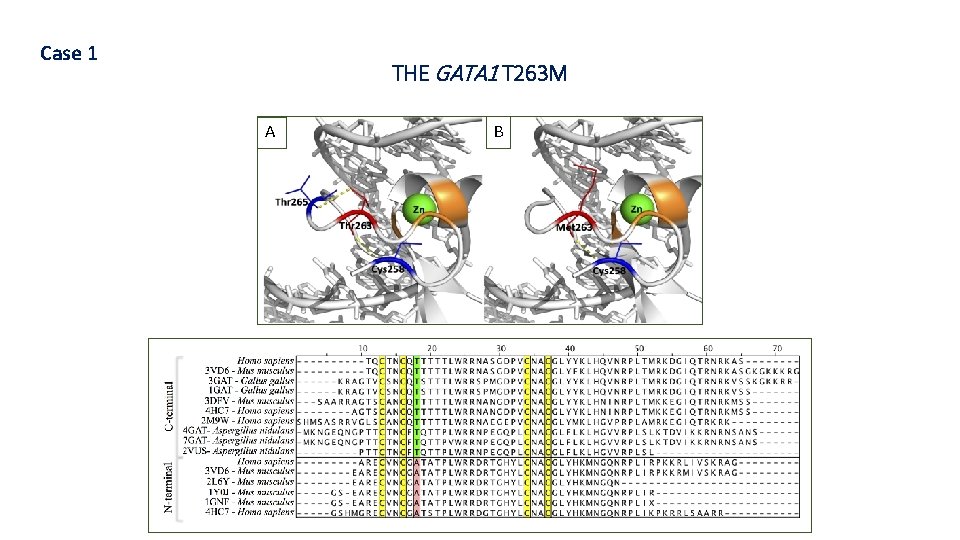
Case 1 THE GATA 1 T 263 M A B

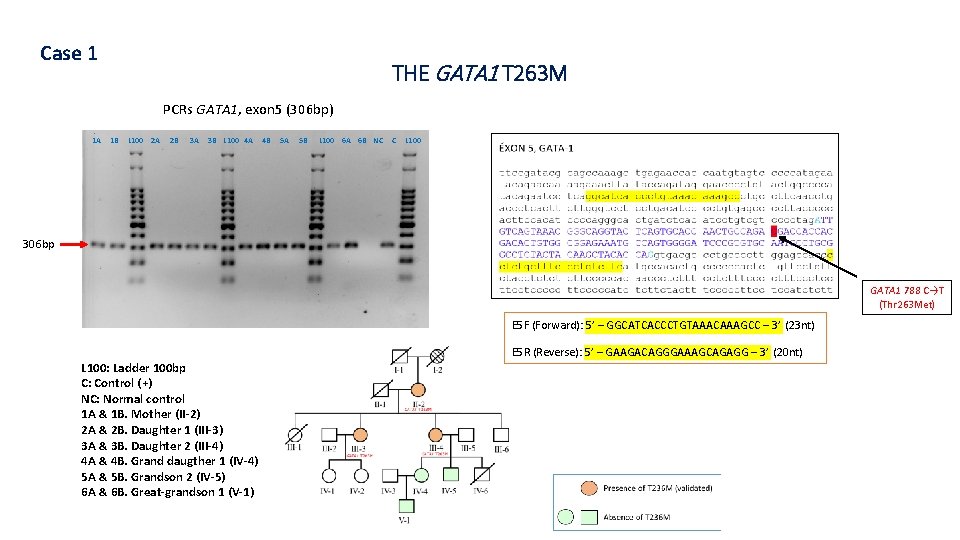
Case 1 THE GATA 1 T 263 M PCRs GATA 1, exon 5 (306 bp) 1 A 1 B L 100 2 A 2 B 3 A 3 B L 100 4 A 4 B 5 A 5 B L 100 6 A 6 B NC C L 100 306 bp GATA 1 788 C→T (Thr 263 Met) E 5 F (Forward): 5’ – GGCATCACCCTGTAAACAAAGCC – 3’ (23 nt) E 5 R (Reverse): 5’ – GAAGACAGGGAAAGCAGAGG – 3’ (20 nt) L 100: Ladder 100 bp C: Control (+) NC: Normal control 1 A & 1 B. Mother (II-2) 2 A & 2 B. Daughter 1 (III-3) 3 A & 3 B. Daughter 2 (III-4) 4 A & 4 B. Grand daugther 1 (IV-4) 5 A & 5 B. Grandson 2 (IV-5) 6 A & 6 B. Great-grandson 1 (V-1)

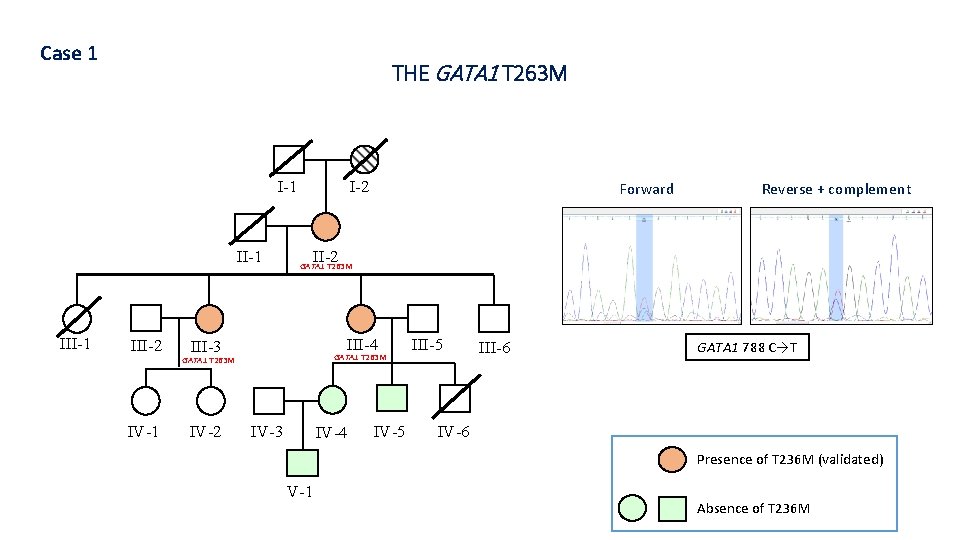
Case 1 THE GATA 1 T 263 M I-1 III-1 III-2 IV-1 I-2 Reverse + complement II-2 GATA 1 T 263 M III-4 III-3 GATA 1 T 263 M IV-2 Forward IV-3 IV-4 IV-5 III-6 GATA 1 788 C→T IV-6 Presence of T 236 M (validated) V-1 Absence of T 236 M

GATA 1 GENE § GATA 1 is one of the transcription factors regulating the development of the precursor bypotential erythrocytes and megakaryocyte (MEP); § Mutations in this gene may be presented in: § Familial dyserythropoietic anemia; (Nichols et al. Nat Genet 2000) § Thrombocytopenia X-linked; (Freson et al. Blood, 2001) § Nearly all cases of transient myeloproliferative disease (TMD) and acute megakaryoblastic leukemia in Down syndrome (DSAMKL); (Harigae et al. Blood, 2005) Crispino; Weiss. Blood, 2014. § AMKL cases not Down syndrome. (Harigae et al. Blood, 2005)

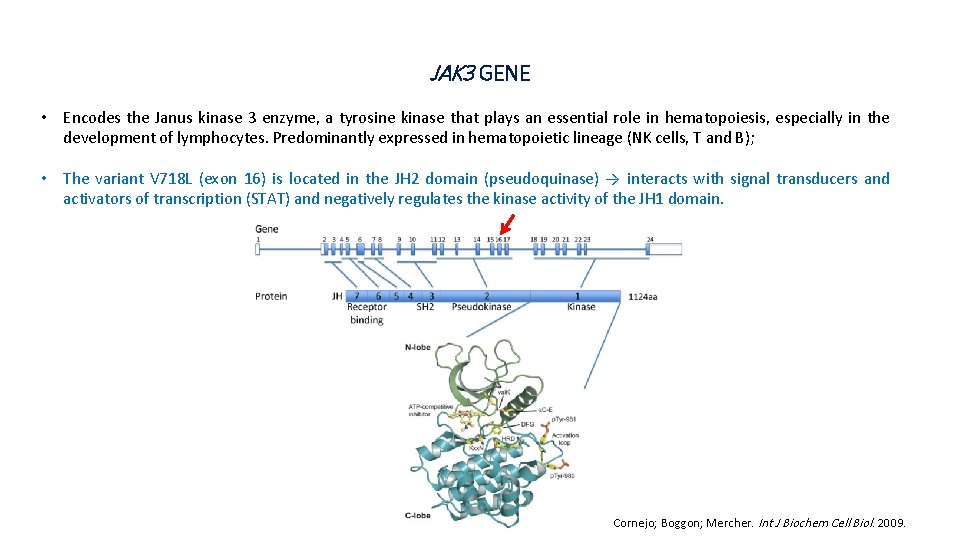
JAK 3 GENE • Encodes the Janus kinase 3 enzyme, a tyrosine kinase that plays an essential role in hematopoiesis, especially in the development of lymphocytes. Predominantly expressed in hematopoietic lineage (NK cells, T and B); • The variant V 718 L (exon 16) is located in the JH 2 domain (pseudoquinase) → interacts with signal transducers and activators of transcription (STAT) and negatively regulates the kinase activity of the JH 1 domain. Cornejo; Boggon; Mercher. Int J Biochem Cell Biol. 2009.

CYP 3 A 5, CYP 2 A 4 and CYP 2 B 6 q The CYP superfamily comprises phase I detoxification enzymes that catalyze the biotransformation of many endogenous and exogenous genotoxic compounds; (Tamási et al. Inflamm Res. 2003) q Many of these enzymes are polymorphic → some polymorphisms are able to alter the enzymatic activity: q modifying the response to treatment and drug resistance; q increasing the risk of developing cancer; q Some SNPs in genes CYP (p. Ex. CYP 2 B 6, CYP 2 B 6 and G 516 T G 15631 T) have been associated with increased susceptibility to chromosomal abnormalities [-7 / Del (7 q) -5 / del (5 q), +8, +21 or t (8; 21)] in AML, ALL and MDS; (Daraki et al. PLo. S One, 2014) q Although CYP 2 B 6 Thr 67 Met variant has not previously been associated with chromosomal changes, the BM cytogenetics of the oldest sister (III-3), where the variant was detected, showed monosomy of chromosome 7 in 35% of analyzed metaphases.

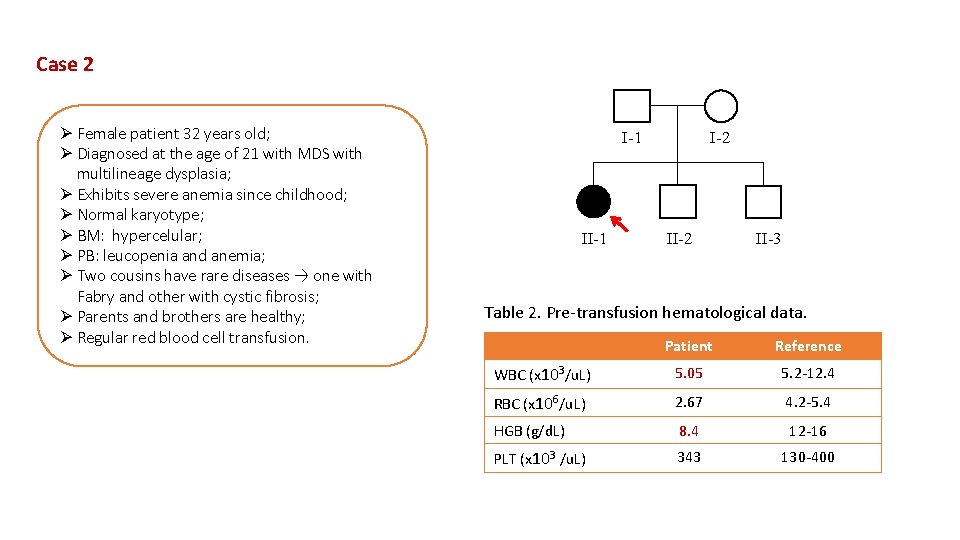
Case 2 Ø Female patient 32 years old; Ø Diagnosed at the age of 21 with MDS with multilineage dysplasia; Ø Exhibits severe anemia since childhood; Ø Normal karyotype; Ø BM: hypercelular; Ø PB: leucopenia and anemia; Ø Two cousins have rare diseases → one with Fabry and other with cystic fibrosis; Ø Parents and brothers are healthy; Ø Regular red blood cell transfusion. I-1 I-2 II-3 Table 2. Pre-transfusion hematological data. Patient Reference WBC (x 103/u. L) 5. 05 5. 2 -12. 4 RBC (x 106/u. L) 2. 67 4. 2 -5. 4 HGB (g/d. L) 8. 4 12 -16 PLT (x 103 /u. L) 343 130 -400

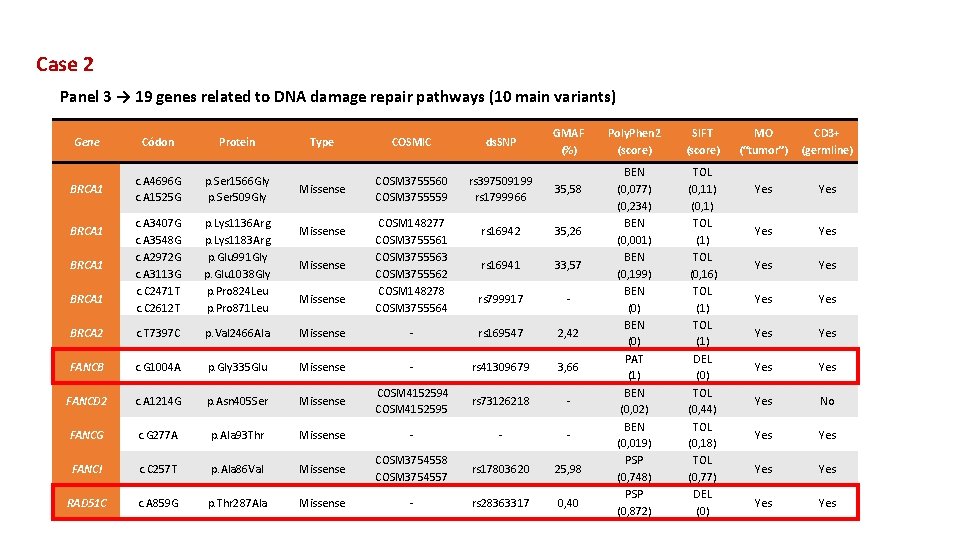
Case 2 Panel 3 → 19 genes related to DNA damage repair pathways (10 main variants) Gene Códon Protein BRCA 1 c. A 4696 G c. A 1525 G p. Ser 1566 Gly p. Ser 509 Gly c. A 3407 G c. A 3548 G c. A 2972 G c. A 3113 G c. C 2471 T c. C 2612 T p. Lys 1136 Arg p. Lys 1183 Arg p. Glu 991 Gly p. Glu 1038 Gly p. Pro 824 Leu p. Pro 871 Leu BRCA 2 c. T 7397 C p. Val 2466 Ala Missense FANCB c. G 1004 A p. Gly 335 Glu FANCD 2 c. A 1214 G FANCG GMAF (%) Type COSMIC ds. SNP Missense COSM 3755560 COSM 3755559 rs 397509199 rs 1799966 35, 58 rs 16942 35, 26 rs 16941 33, 57 rs 799917 - - rs 169547 2, 42 Missense - rs 41309679 3, 66 p. Asn 405 Ser Missense COSM 4152594 COSM 4152595 rs 73126218 - c. G 277 A p. Ala 93 Thr Missense - - - FANCI c. C 257 T p. Ala 86 Val Missense COSM 3754558 COSM 3754557 rs 17803620 25, 98 RAD 51 C c. A 859 G p. Thr 287 Ala Missense - rs 28363317 0, 40 BRCA 1 Missense COSM 148277 COSM 3755561 COSM 3755563 COSM 3755562 COSM 148278 COSM 3755564 Poly. Phen 2 (score) SIFT (score) BEN (0, 077) (0, 234) BEN (0, 001) BEN (0, 199) BEN (0) PAT (1) BEN (0, 02) BEN (0, 019) PSP (0, 748) PSP (0, 872) TOL (0, 11) (0, 1) TOL (0, 16) TOL (1) DEL (0) TOL (0, 44) TOL (0, 18) TOL (0, 77) DEL (0) MO (“tumor”) CD 3+ (germline) Yes Yes Yes Yes No Yes Yes Yes

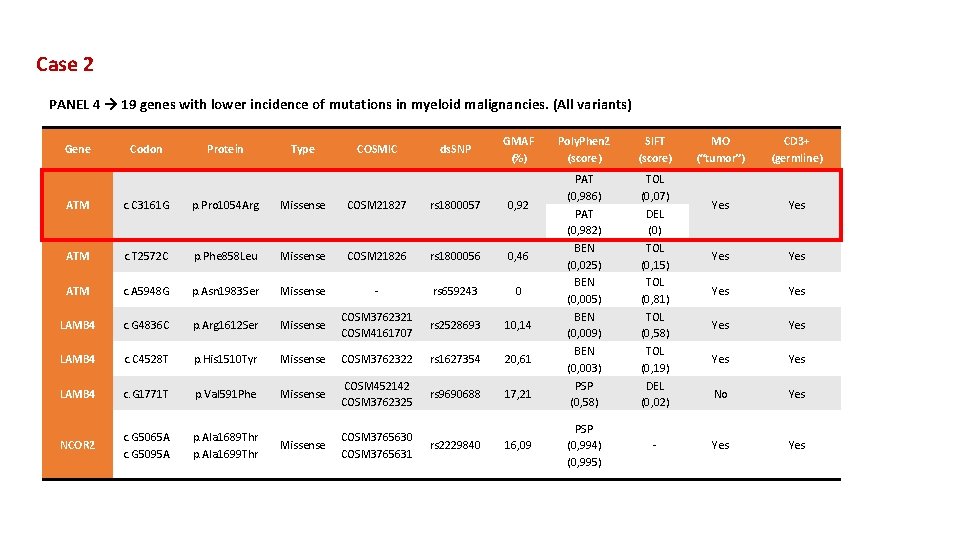
Case 2 PANEL 4 → 19 genes with lower incidence of mutations in myeloid malignancies. (All variants) Gene Codon Protein Type COSMIC ds. SNP GMAF (%) ATM c. C 3161 G p. Pro 1054 Arg Missense COSM 21827 rs 1800057 0, 92 ATM c. T 2572 C p. Phe 858 Leu Missense COSM 21826 rs 1800056 0, 46 ATM c. A 5948 G p. Asn 1983 Ser Missense - rs 659243 0 LAMB 4 c. G 4836 C p. Arg 1612 Ser Missense COSM 3762321 COSM 4161707 rs 2528693 10, 14 LAMB 4 c. C 4528 T p. His 1510 Tyr Missense COSM 3762322 rs 1627354 20, 61 LAMB 4 c. G 1771 T p. Val 591 Phe Missense COSM 452142 COSM 3762325 rs 9690688 17, 21 NCOR 2 c. G 5065 A c. G 5095 A p. Ala 1689 Thr p. Ala 1699 Thr Missense COSM 3765630 COSM 3765631 rs 2229840 16, 09 Poly. Phen 2 (score) SIFT (score) PAT (0, 986) PAT (0, 982) BEN (0, 025) BEN (0, 009) BEN (0, 003) PSP (0, 58) TOL (0, 07) DEL (0) TOL (0, 15) TOL (0, 81) TOL (0, 58) TOL (0, 19) DEL (0, 02) PSP (0, 994) (0, 995) - MO (“tumor”) CD 3+ (germline) Yes Yes Yes No Yes Yes

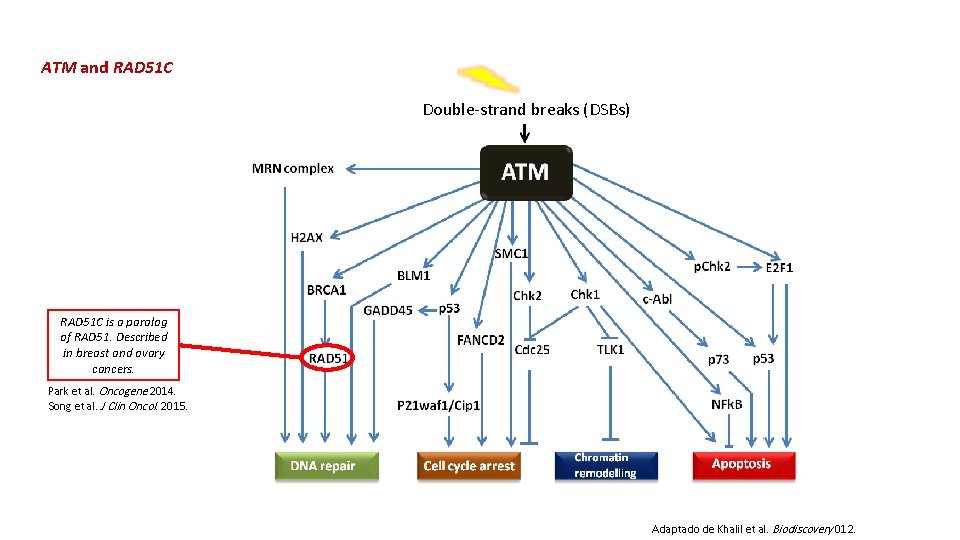
ATM and RAD 51 C Double-strand breaks (DSBs) RAD 51 C is a paralog of RAD 51. Described in breast and ovary cancers. Park et al. Oncogene 2014. Song et al. J Clin Oncol. 2015. Adaptado de Khalil et al. Biodiscovery 012.

ATM, RAD 51 C and FANCB q The missense ATM Pro 1054 Arg variant has previously been associated with an increased risk of prostate cancer and differentiated thyroid carcinoma, but had not yet been reported in hematologic malignancies. (Henríquez-Hernández et al. PLo. S One. 2013) (Xu et al. J Clin Endocrinol Metab. 2012) q The variant RAD 51 C Thr 287 Ala has been widely reported in the literature on its occurrence as in patients with breast and or ovarian cancer, but has not been described in myeloid neoplasms yet. (Meindl et al. Nat Genet, 2010) (Romero et al. Breast Cancer Res Treat. 2011) (Pelttari et al. Hum Mol Genet 2011) (Akbari et al. Breast Cancer Res 2010). q Mutations in FANCB are involved in very high risk of developing acute myeloid leukemia and myelodysplastic syndrome and extremely high risk for particular solid tumors. (Alter. Clin Haematol. 2014)

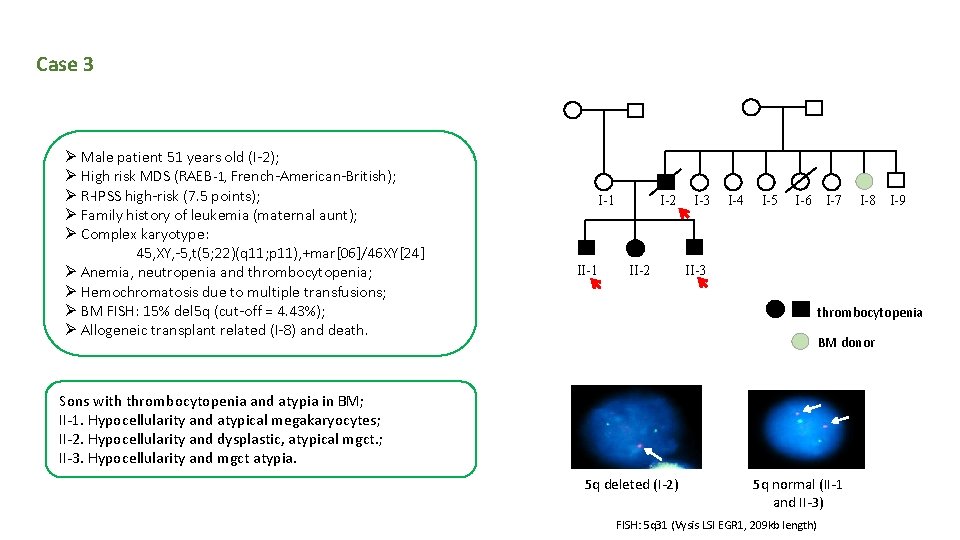
Case 3 Ø Male patient 51 years old (I-2); Ø High risk MDS (RAEB-1, French-American-British); Ø R-IPSS high-risk (7. 5 points); Ø Family history of leukemia (maternal aunt); Ø Complex karyotype: 45, XY, -5, t(5; 22)(q 11; p 11), +mar[06]/46 XY[24] Ø Anemia, neutropenia and thrombocytopenia; Ø Hemochromatosis due to multiple transfusions; Ø BM FISH: 15% del 5 q (cut-off = 4. 43%); Ø Allogeneic transplant related (I-8) and death. I-1 I-2 II-2 I-3 I-4 I-5 I-6 I-7 I-8 I-9 II-3 thrombocytopenia BM donor Sons with thrombocytopenia and atypia in BM; II-1. Hypocellularity and atypical megakaryocytes; II-2. Hypocellularity and dysplastic, atypical mgct. ; II-3. Hypocellularity and mgct atypia. 5 q deleted (I-2) 5 q normal (II-1 and II-3) FISH: 5 q 31 (Vysis LSI EGR 1, 209 kb length)

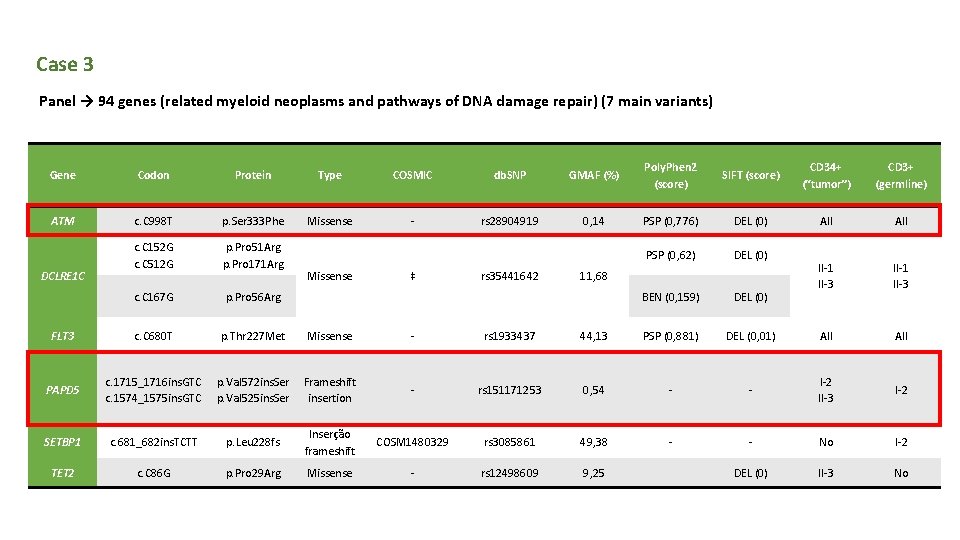
Case 3 Panel → 94 genes (related myeloid neoplasms and pathways of DNA damage repair) (7 main variants) Gene Codon Protein Type COSMIC db. SNP GMAF (%) Poly. Phen 2 (score) SIFT (score) CD 34+ (“tumor”) CD 3+ (germline) ATM c. C 998 T p. Ser 333 Phe Missense - rs 28904919 0, 14 PSP (0, 776) DEL (0) All c. C 152 G c. C 512 G p. Pro 51 Arg p. Pro 171 Arg PSP (0, 62) DEL (0) c. C 167 G p. Pro 56 Arg BEN (0, 159) DEL (0) II-1 II-3 FLT 3 c. C 680 T p. Thr 227 Met Missense - rs 1933437 44, 13 PSP (0, 881) DEL (0, 01) All PAPD 5 c. 1715_1716 ins. GTC c. 1574_1575 ins. GTC p. Val 572 ins. Ser p. Val 525 ins. Ser Frameshift insertion - rs 151171253 0, 54 - - I-2 II-3 I-2 SETBP 1 c. 681_682 ins. TCTT p. Leu 228 fs Inserção frameshift COSM 1480329 rs 3085861 49, 38 - - No I-2 TET 2 c. C 86 G p. Pro 29 Arg Missense - rs 12498609 9, 25 DEL (0) II-3 No DCLRE 1 C Missense ‡ rs 35441642 11, 68

PAPD 5 q PAPD 5 → mechanisms of post-transcriptional control activity → has poly (A) RNA polymerase and is involved in the degradation of m. RNAs from histones uridylation dependent. q PAPD 5 Val 572 ins. Ser has not been reported in myeloid neoplasms.

Conclusion • Description of novel mutations causing bone marrow malignancies • Important for physiopathology understanding and therapeutic development

Fabiola Traina, MD, Ph. D Daniela Basseres, Ph. D Carolina Bigarella, Ph. D Luciene Borges, Ph. D Patricia Favaro Ph. D Joao A Machado-Neto MSc Juliana Xavier, Bs. C Mariana Lazarini Ph. D Mariana Baratti Ph. D Adriana SS Duarte Ph. D Leticia F. Archangelo Ph. D Paula M Campos MD Bruno Benites MD Karin Barcellos, Ph. D Matheus Arouca, MSC Moises Alves, MSC Tereza SI Sales, Bs. C Bruna Palodeto Flavia Correcher Rita Melo Fernanda Roversi Genomics Facility of UNICAMP Acknowledgments
 Campinas 2005 accident
Campinas 2005 accident Vigiagro viracopos
Vigiagro viracopos Centro de saúde jardim vista alegre
Centro de saúde jardim vista alegre Cmdca campinas
Cmdca campinas Mercer medical library
Mercer medical library Multlab
Multlab Uel
Uel Modle ufv
Modle ufv O que é paais unicamp
O que é paais unicamp Acervus unicamp
Acervus unicamp Fcm unicamp
Fcm unicamp Mc102 unicamp
Mc102 unicamp Terrômetro
Terrômetro Cad unicamp
Cad unicamp Unicamp reproduzimos abaixo a chamada
Unicamp reproduzimos abaixo a chamada Nelson fonseca unicamp
Nelson fonseca unicamp Dgrh unicamp vida funcional
Dgrh unicamp vida funcional Faculdade de tecnologia da unicamp
Faculdade de tecnologia da unicamp Ufrgs diamante e grafite sao variedades
Ufrgs diamante e grafite sao variedades Salem state msw
Salem state msw University of medicine and pharmacy timisoara
University of medicine and pharmacy timisoara Hubert kairuki memorial university faculty of medicine
Hubert kairuki memorial university faculty of medicine Semmelweis
Semmelweis Lincoln memorial university college of veterinary medicine
Lincoln memorial university college of veterinary medicine King saud university college of medicine
King saud university college of medicine King saud university college of medicine
King saud university college of medicine King saud university college of medicine
King saud university college of medicine Applied medical sciences
Applied medical sciences University of wisconsin integrative medicine anxiety
University of wisconsin integrative medicine anxiety




















































