Stat Quest PCA Clearly Explained by Joshua Starmer

Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
quest Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

quest Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

Stat. Quest!!! Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
Stat. Quest!!! Principal Component Analysis (PCA) Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

Let’s start with an example of Principal Component Analysis (PCA) in action… Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
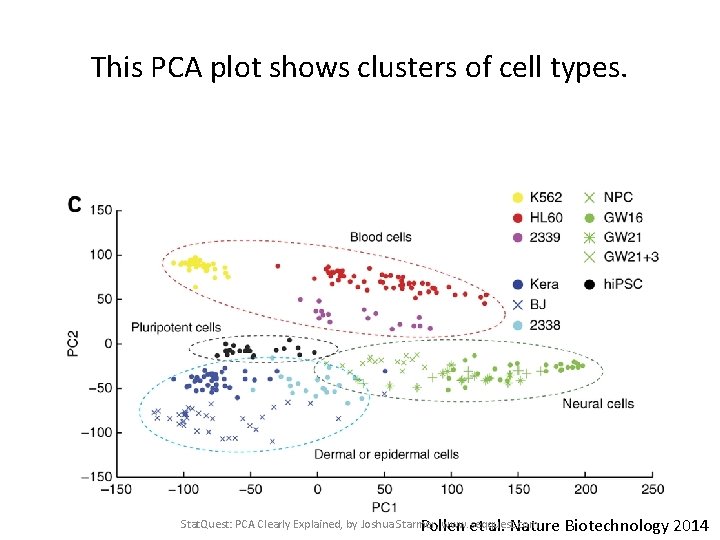
This PCA plot shows clusters of cell types. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com Pollen et al. Nature Biotechnology 2014
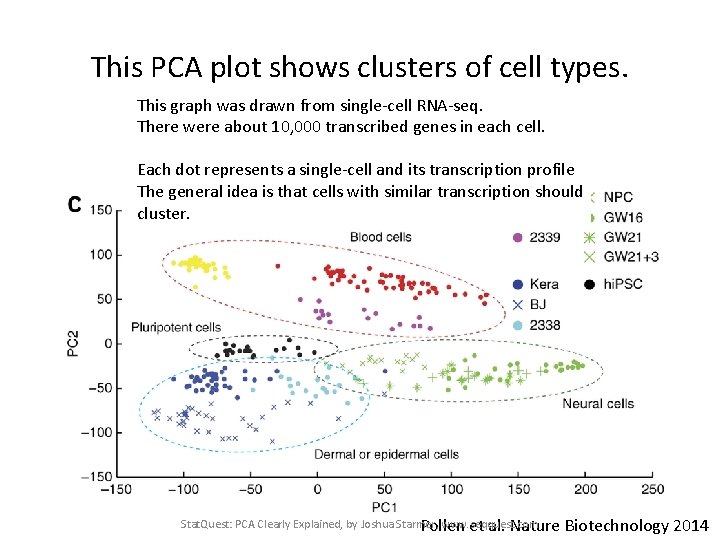
This PCA plot shows clusters of cell types. This graph was drawn from single-cell RNA-seq. There were about 10, 000 transcribed genes in each cell. Each dot represents a single-cell and its transcription profile The general idea is that cells with similar transcription should cluster. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com Pollen et al. Nature Biotechnology 2014
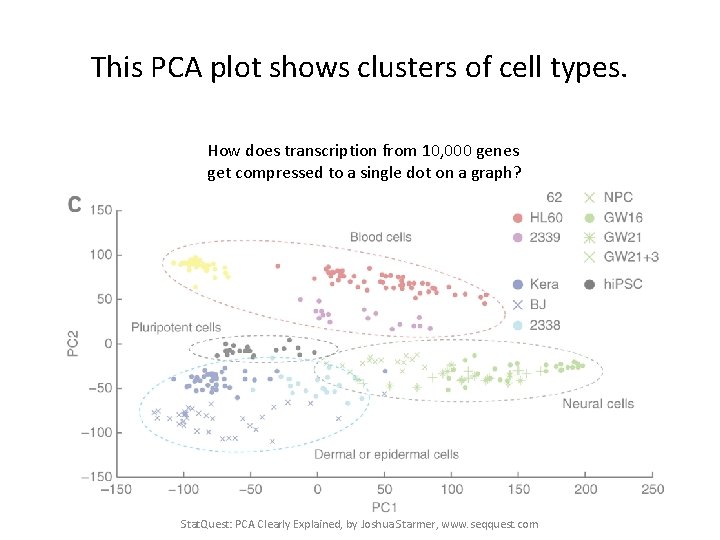
This PCA plot shows clusters of cell types. How does transcription from 10, 000 genes get compressed to a single dot on a graph? Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
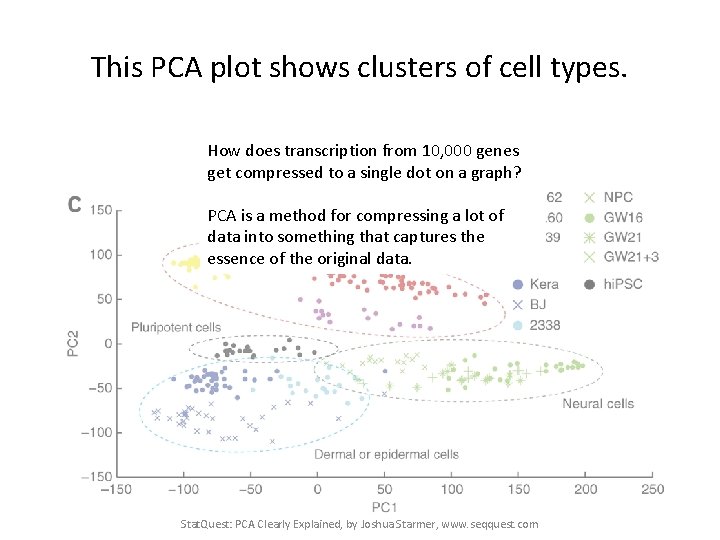
This PCA plot shows clusters of cell types. How does transcription from 10, 000 genes get compressed to a single dot on a graph? PCA is a method for compressing a lot of data into something that captures the essence of the original data. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
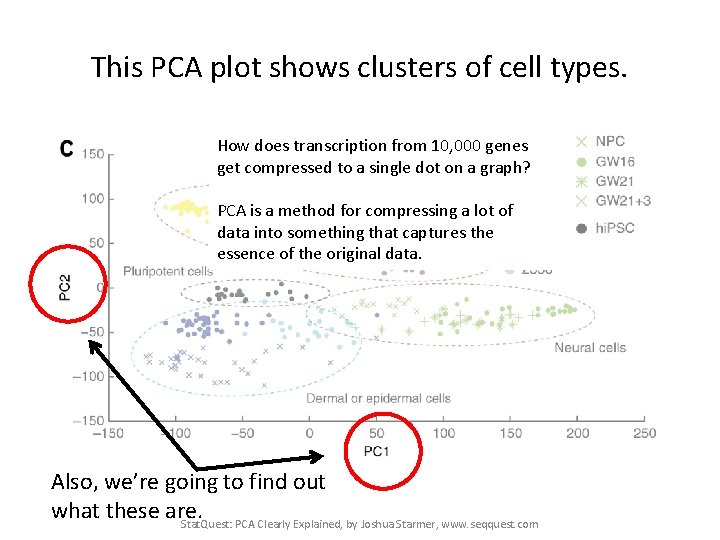
This PCA plot shows clusters of cell types. How does transcription from 10, 000 genes get compressed to a single dot on a graph? PCA is a method for compressing a lot of data into something that captures the essence of the original data. Also, we’re going to find out what these are. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

Background: An Introduction to Dimensions Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

Background: An Introduction to Dimensions • This is going to seem very, very simple. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

Background: An Introduction to Dimensions • This is going to seem very, very simple. • Just hang in there, you’ll be glad we did this. – It will keep your head from exploding. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
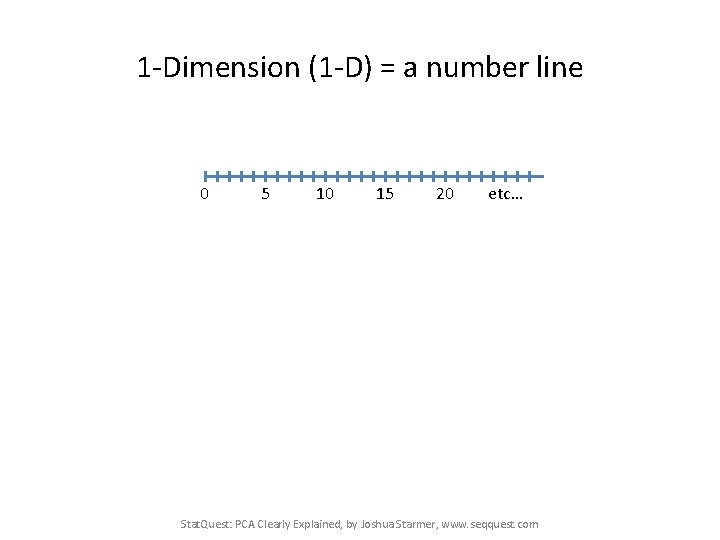
1 -Dimension (1 -D) = a number line 0 5 10 15 20 etc… Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

1 -Dimension (1 -D) = a number line 0 5 10 15 20 etc… A pretend RNA-seq data set for a single cell: Gene: A B C … Reads: 10 0 14 … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
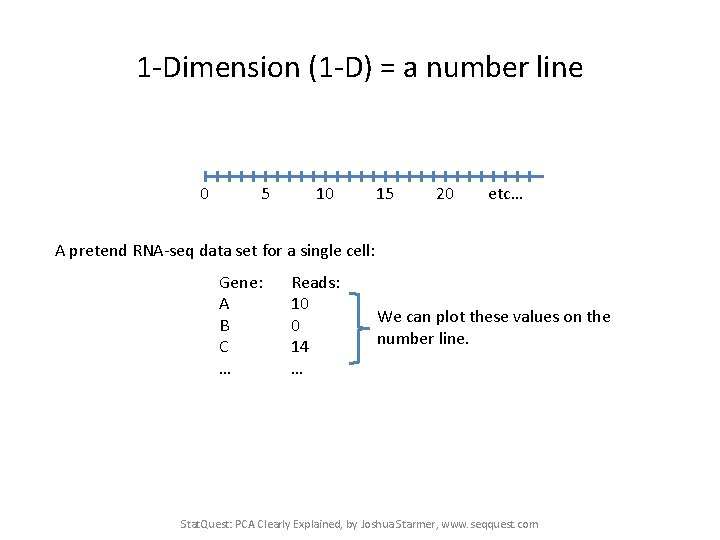
1 -Dimension (1 -D) = a number line 0 5 10 15 20 etc… A pretend RNA-seq data set for a single cell: Gene: A B C … Reads: 10 0 14 … We can plot these values on the number line. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

1 -Dimension (1 -D) = a number line A 0 5 10 15 20 etc… A pretend RNA-seq data set for a single cell: Gene: A B C … Reads: 10 0 14 … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
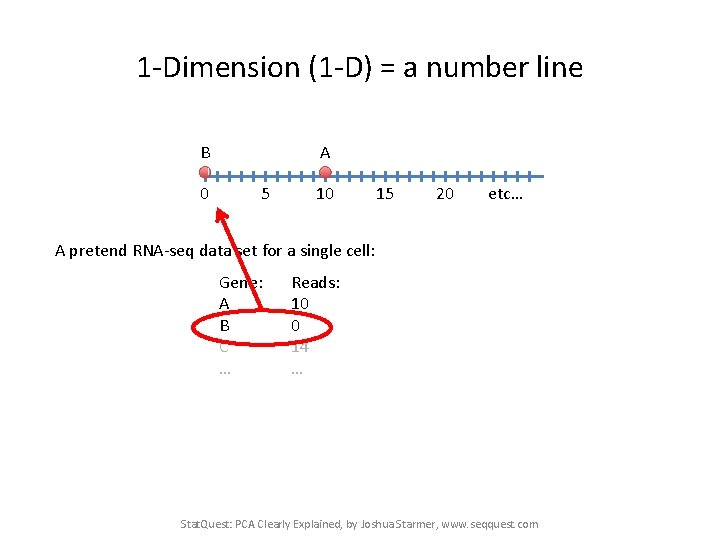
1 -Dimension (1 -D) = a number line B 0 A 5 10 15 20 etc… A pretend RNA-seq data set for a single cell: Gene: A B C … Reads: 10 0 14 … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

1 -Dimension (1 -D) = a number line B 0 A 5 C 10 15 20 etc… A pretend RNA-seq data set for a single cell: Gene: A B C … Reads: 10 0 14 … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

1 -Dimension (1 -D) = a number line B 0 A 5 C 10 15 20 etc… A pretend RNA-seq data set for a single cell: Gene: A B C … Low Reads: 10 0 14 … If we plotted all genes, we might see something like this High A uniform distribution of transcripts Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
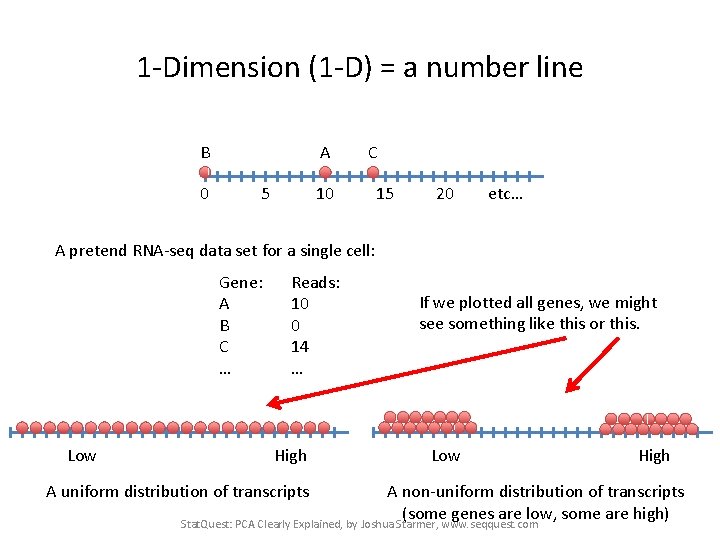
1 -Dimension (1 -D) = a number line B 0 A 5 C 10 15 20 etc… A pretend RNA-seq data set for a single cell: Gene: A B C … Low Reads: 10 0 14 … High A uniform distribution of transcripts If we plotted all genes, we might see something like this or this. Low High A non-uniform distribution of transcripts (some genes are low, some are high) Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
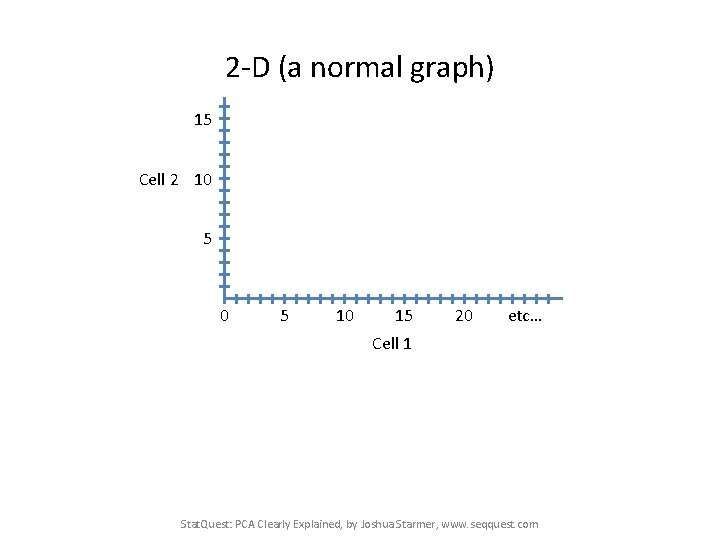
2 -D (a normal graph) 15 Cell 2 10 5 10 15 20 etc… Cell 1 Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
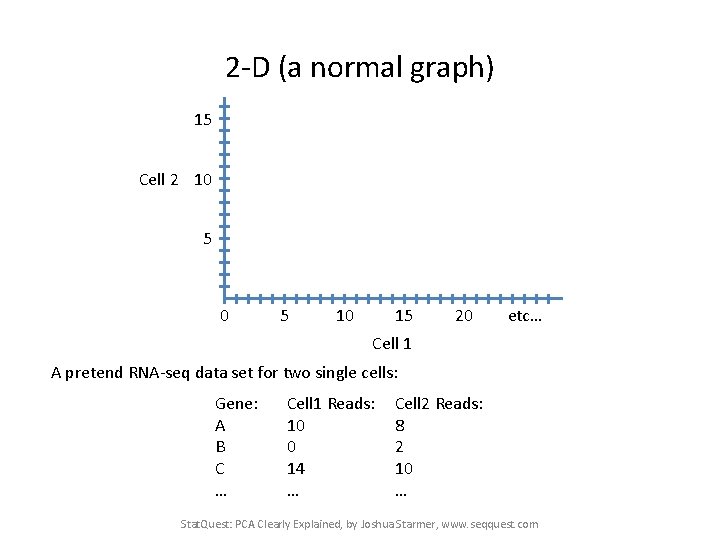
2 -D (a normal graph) 15 Cell 2 10 5 10 15 20 etc… Cell 1 A pretend RNA-seq data set for two single cells: Gene: A B C … Cell 1 Reads: 10 0 14 … Cell 2 Reads: 8 2 10 … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
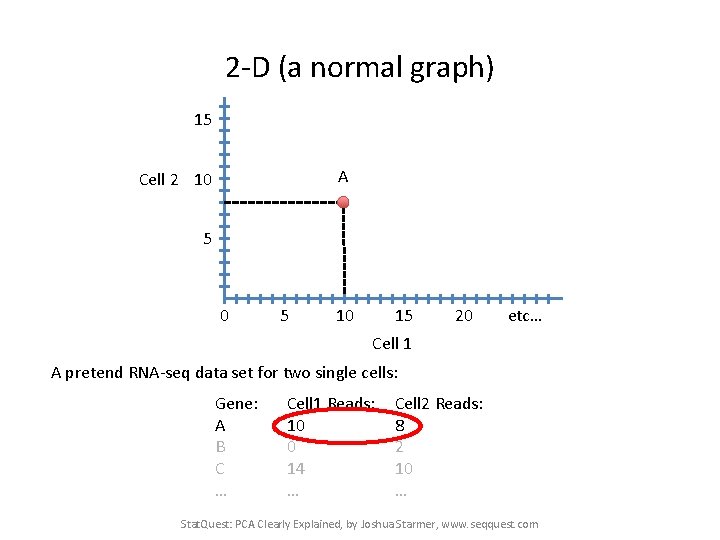
2 -D (a normal graph) 15 A Cell 2 10 5 10 15 20 etc… Cell 1 A pretend RNA-seq data set for two single cells: Gene: A B C … Cell 1 Reads: 10 0 14 … Cell 2 Reads: 8 2 10 … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
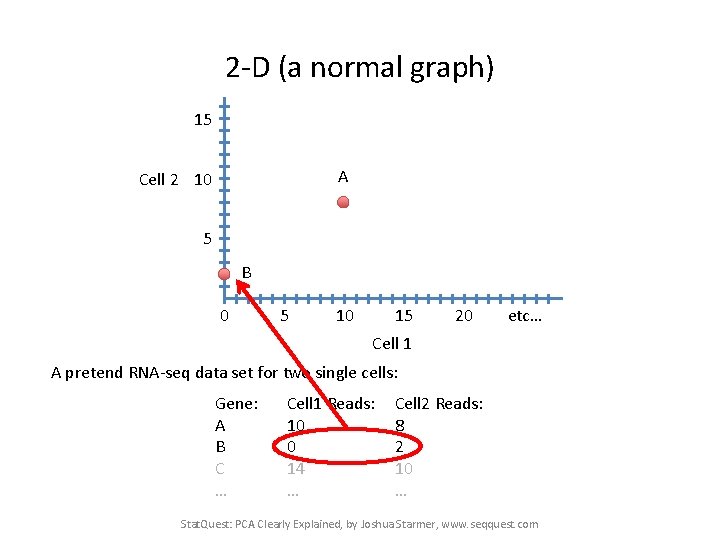
2 -D (a normal graph) 15 A Cell 2 10 5 B 0 5 10 15 20 etc… Cell 1 A pretend RNA-seq data set for two single cells: Gene: A B C … Cell 1 Reads: 10 0 14 … Cell 2 Reads: 8 2 10 … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
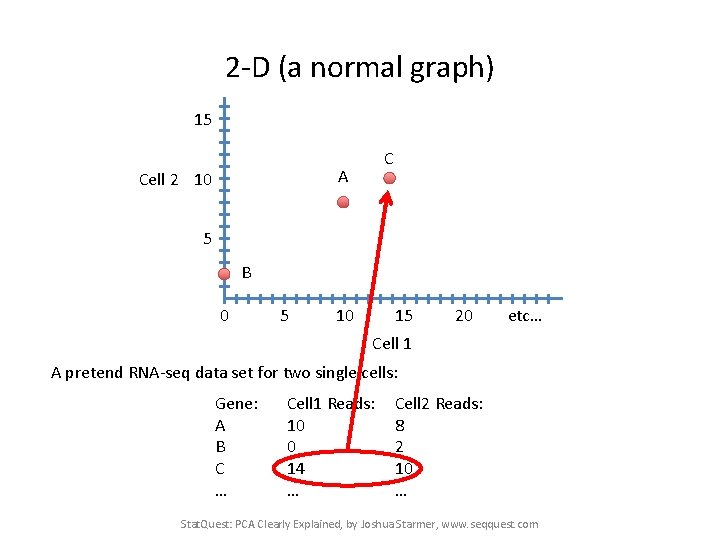
2 -D (a normal graph) 15 C A Cell 2 10 5 B 0 5 10 15 20 etc… Cell 1 A pretend RNA-seq data set for two single cells: Gene: A B C … Cell 1 Reads: 10 0 14 … Cell 2 Reads: 8 2 10 … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
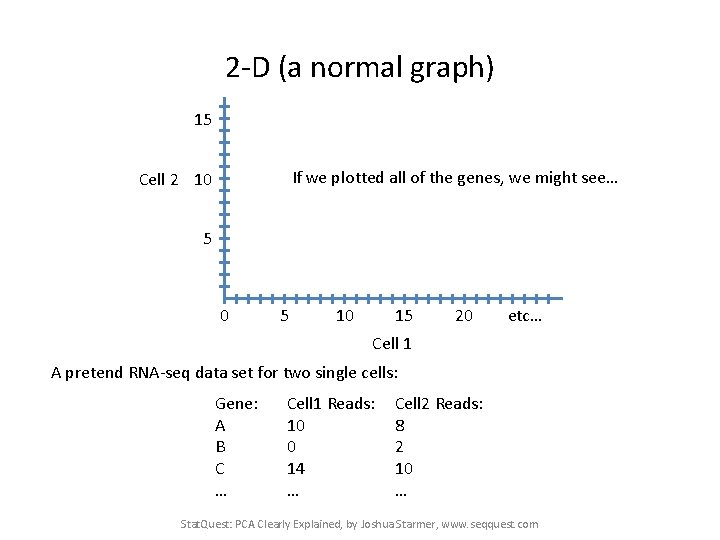
2 -D (a normal graph) 15 If we plotted all of the genes, we might see… Cell 2 10 5 10 15 20 etc… Cell 1 A pretend RNA-seq data set for two single cells: Gene: A B C … Cell 1 Reads: 10 0 14 … Cell 2 Reads: 8 2 10 … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
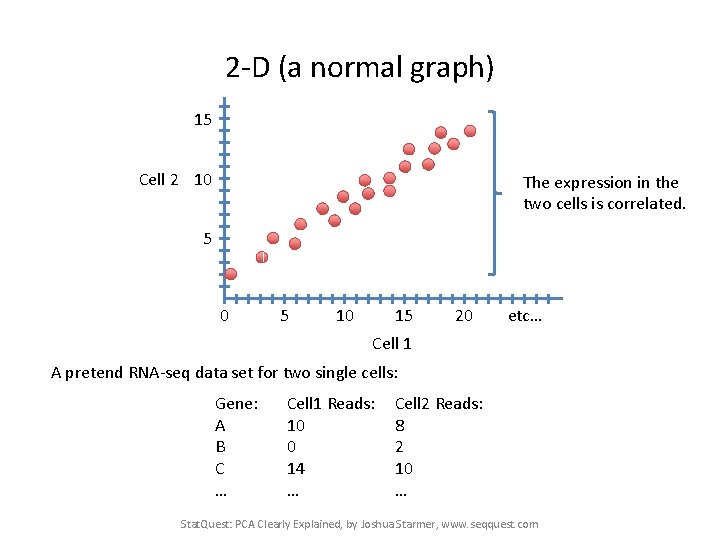
2 -D (a normal graph) 15 Cell 2 10 The expression in the two cells is correlated. 5 0 5 10 15 20 etc… Cell 1 A pretend RNA-seq data set for two single cells: Gene: A B C … Cell 1 Reads: 10 0 14 … Cell 2 Reads: 8 2 10 … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
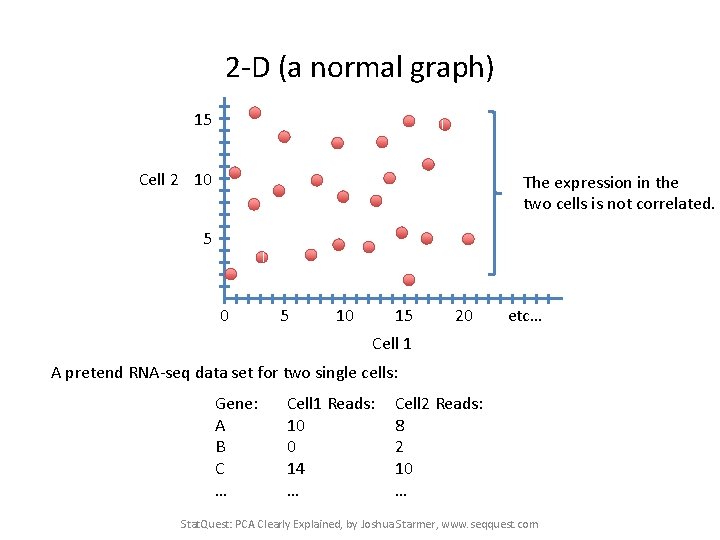
2 -D (a normal graph) 15 Cell 2 10 The expression in the two cells is not correlated. 5 0 5 10 15 20 etc… Cell 1 A pretend RNA-seq data set for two single cells: Gene: A B C … Cell 1 Reads: 10 0 14 … Cell 2 Reads: 8 2 10 … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
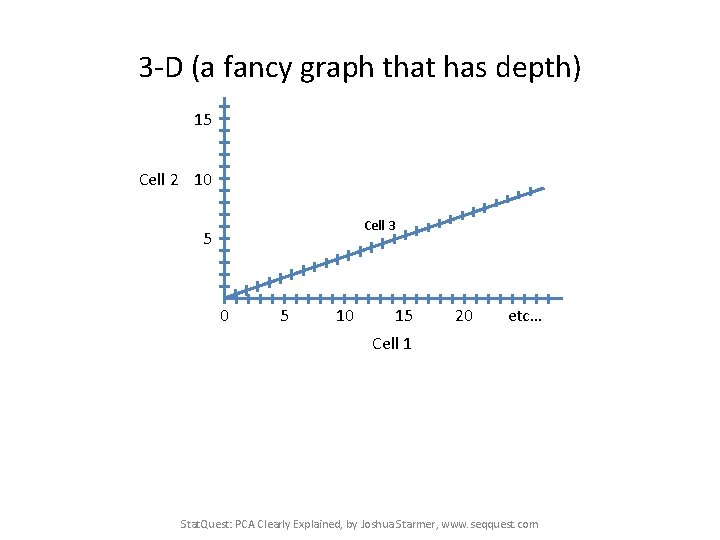
3 -D (a fancy graph that has depth) 15 Cell 2 10 Cell 3 5 0 5 10 15 20 etc… Cell 1 Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
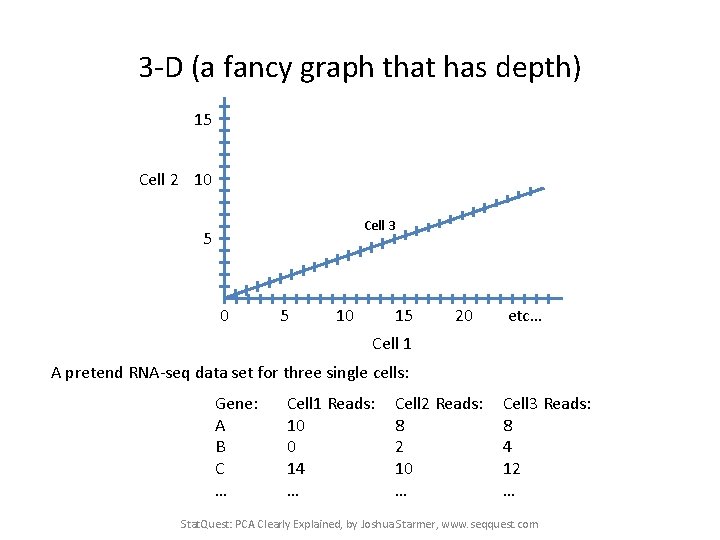
3 -D (a fancy graph that has depth) 15 Cell 2 10 Cell 3 5 0 5 10 15 20 etc… Cell 1 A pretend RNA-seq data set for three single cells: Gene: A B C … Cell 1 Reads: 10 0 14 … Cell 2 Reads: 8 2 10 … Cell 3 Reads: 8 4 12 … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
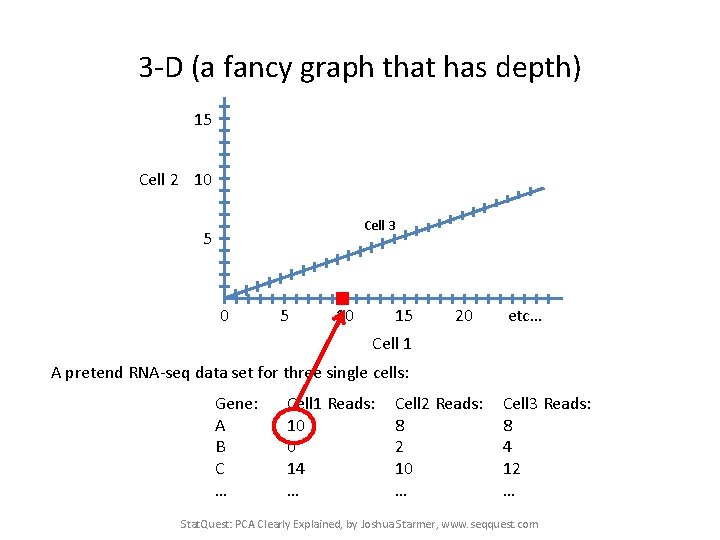
3 -D (a fancy graph that has depth) 15 Cell 2 10 Cell 3 5 0 5 10 15 20 etc… Cell 1 A pretend RNA-seq data set for three single cells: Gene: A B C … Cell 1 Reads: 10 0 14 … Cell 2 Reads: 8 2 10 … Cell 3 Reads: 8 4 12 … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
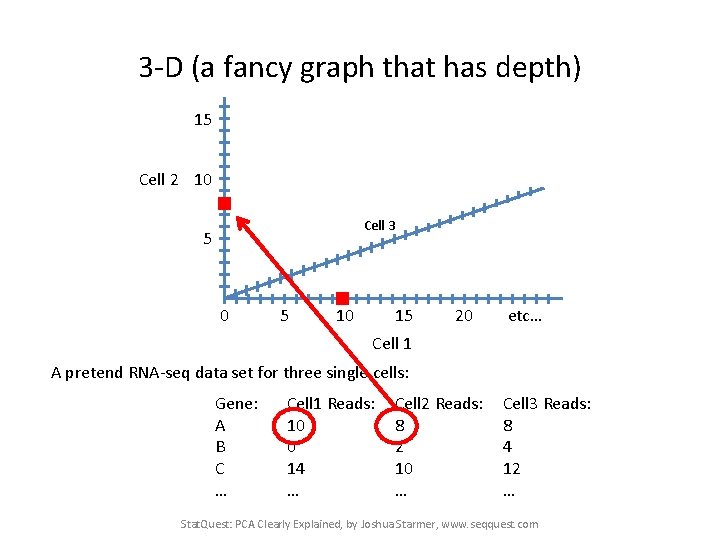
3 -D (a fancy graph that has depth) 15 Cell 2 10 Cell 3 5 0 5 10 15 20 etc… Cell 1 A pretend RNA-seq data set for three single cells: Gene: A B C … Cell 1 Reads: 10 0 14 … Cell 2 Reads: 8 2 10 … Cell 3 Reads: 8 4 12 … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
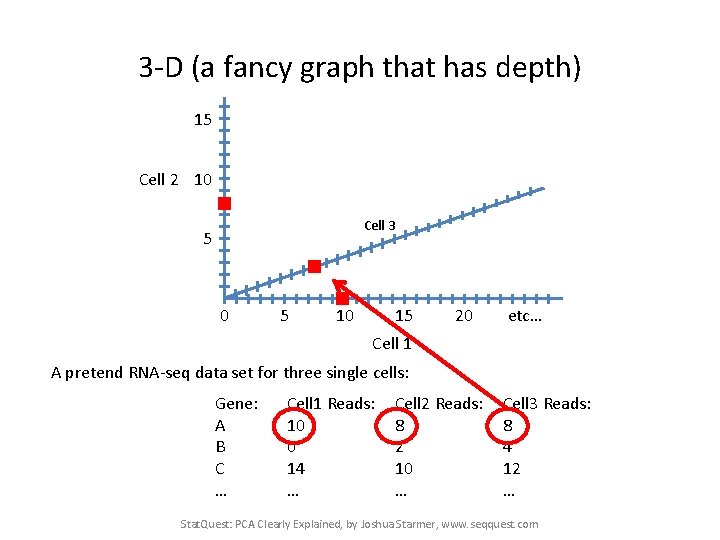
3 -D (a fancy graph that has depth) 15 Cell 2 10 Cell 3 5 0 5 10 15 20 etc… Cell 1 A pretend RNA-seq data set for three single cells: Gene: A B C … Cell 1 Reads: 10 0 14 … Cell 2 Reads: 8 2 10 … Cell 3 Reads: 8 4 12 … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
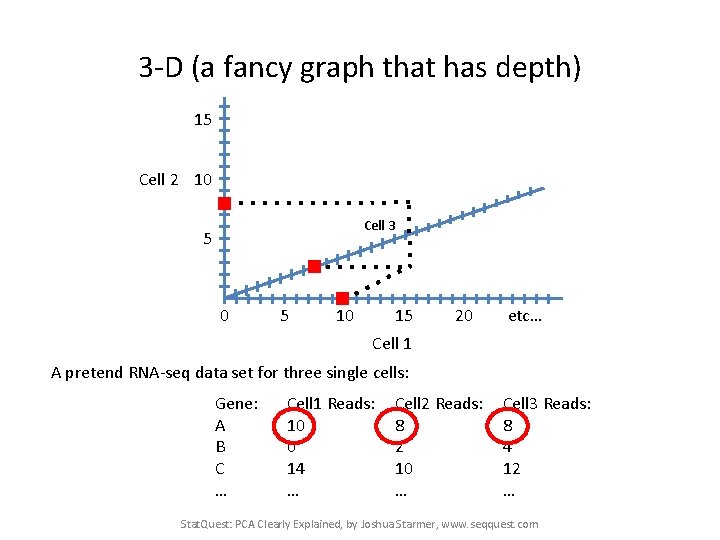
3 -D (a fancy graph that has depth) 15 Cell 2 10 Cell 3 5 0 5 10 15 20 etc… Cell 1 A pretend RNA-seq data set for three single cells: Gene: A B C … Cell 1 Reads: 10 0 14 … Cell 2 Reads: 8 2 10 … Cell 3 Reads: 8 4 12 … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
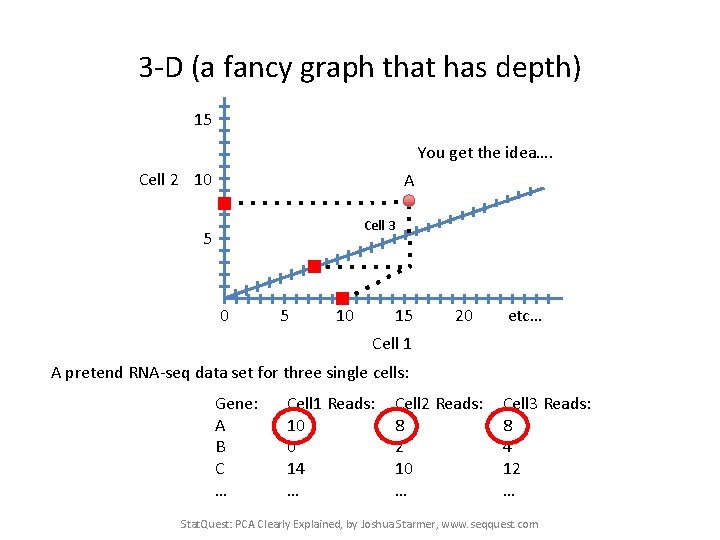
3 -D (a fancy graph that has depth) 15 You get the idea…. Cell 2 10 A Cell 3 5 0 5 10 15 20 etc… Cell 1 A pretend RNA-seq data set for three single cells: Gene: A B C … Cell 1 Reads: 10 0 14 … Cell 2 Reads: 8 2 10 … Cell 3 Reads: 8 4 12 … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

Dimensions So Far… • 1 cell = 1 -D graph (number line) • Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
Dimensions So Far… • 1 cell = 1 -D graph (number line) • 2 cells = 2 -D graph (normal x/y graph) • Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
Dimensions So Far… • 1 cell = 1 -D graph (number line) • 2 cells = 2 -D graph (normal x/y graph) • 3 cells = 3 -D graph (fancy graph with depth) • Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
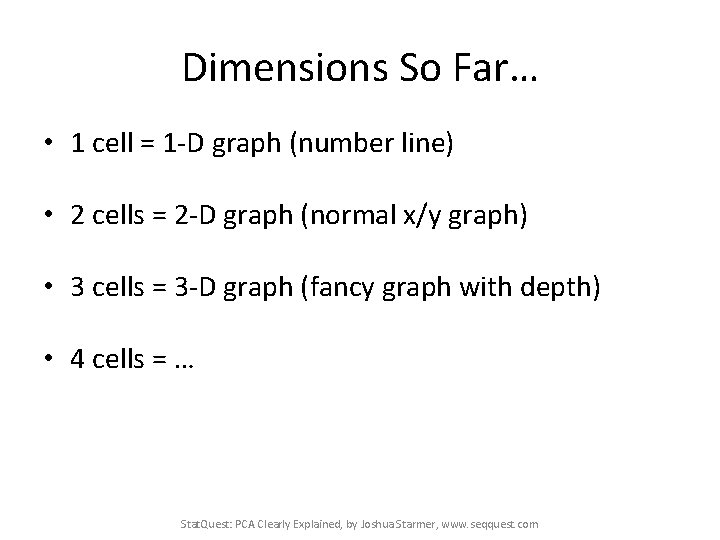
Dimensions So Far… • 1 cell = 1 -D graph (number line) • 2 cells = 2 -D graph (normal x/y graph) • 3 cells = 3 -D graph (fancy graph with depth) • 4 cells = … • Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
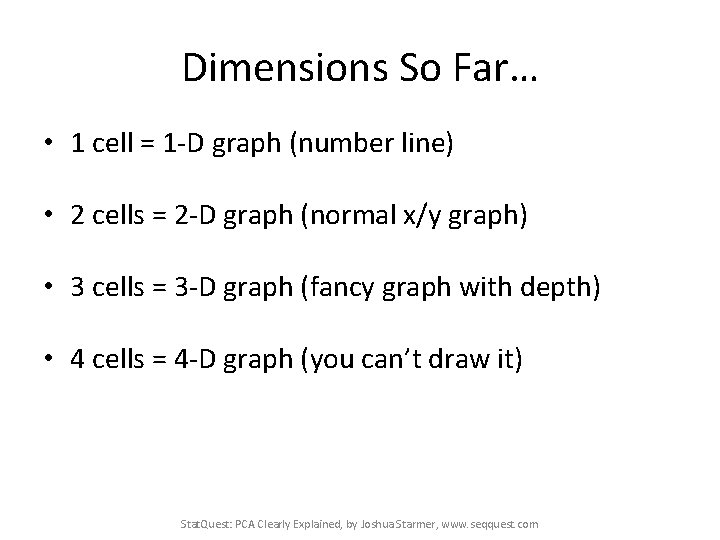
Dimensions So Far… • 1 cell = 1 -D graph (number line) • 2 cells = 2 -D graph (normal x/y graph) • 3 cells = 3 -D graph (fancy graph with depth) • 4 cells = 4 -D graph (you can’t draw it) • Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
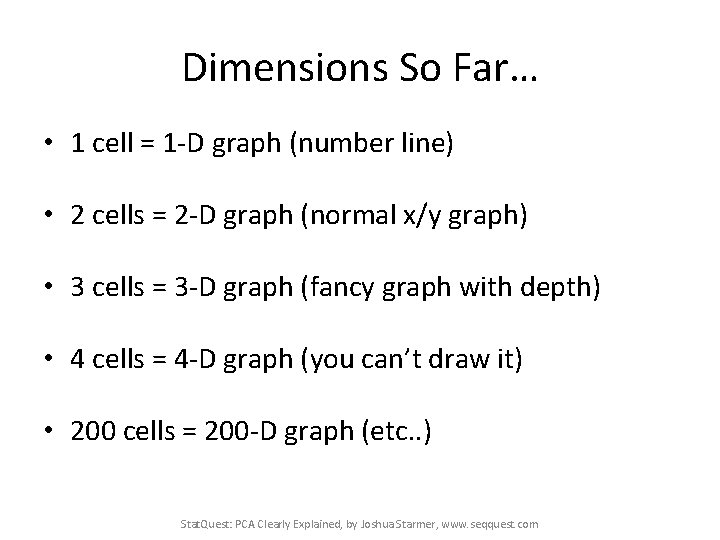
Dimensions So Far… • 1 cell = 1 -D graph (number line) • 2 cells = 2 -D graph (normal x/y graph) • 3 cells = 3 -D graph (fancy graph with depth) • 4 cells = 4 -D graph (you can’t draw it) • 200 cells = 200 -D graph (etc. . ) Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
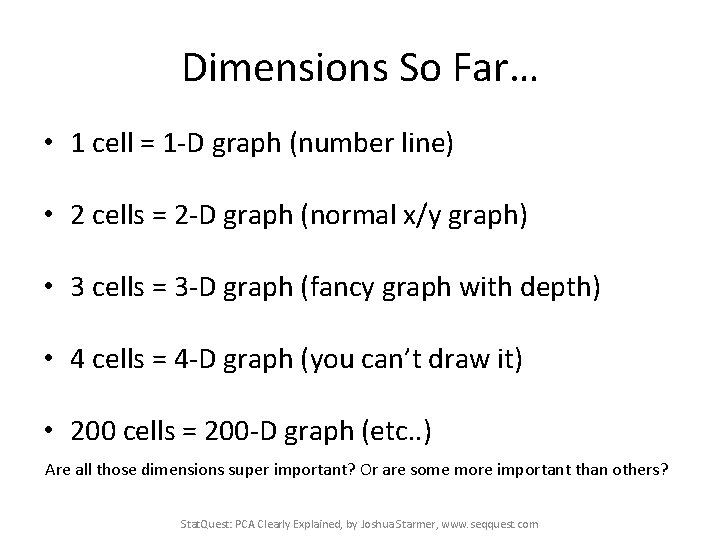
Dimensions So Far… • 1 cell = 1 -D graph (number line) • 2 cells = 2 -D graph (normal x/y graph) • 3 cells = 3 -D graph (fancy graph with depth) • 4 cells = 4 -D graph (you can’t draw it) • 200 cells = 200 -D graph (etc. . ) Are all those dimensions super important? Or are some more important than others? Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

Back to 2 Cells (and 2 Dimensions) Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
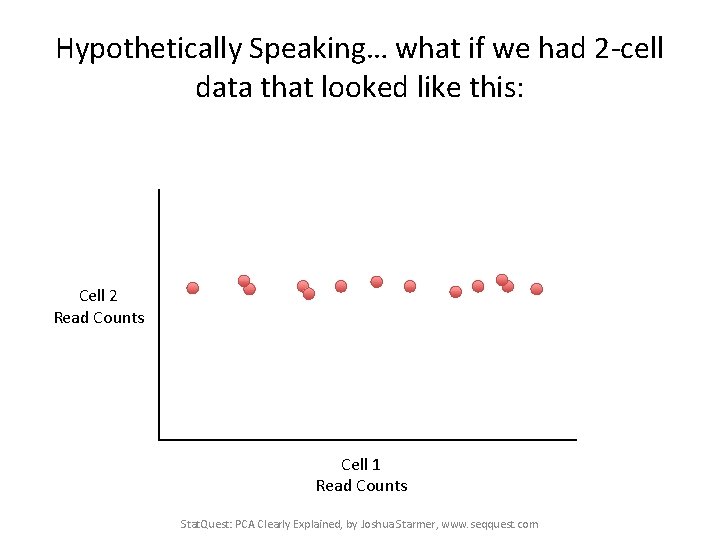
Hypothetically Speaking… what if we had 2 -cell data that looked like this: Cell 2 Read Counts Cell 1 Read Counts Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
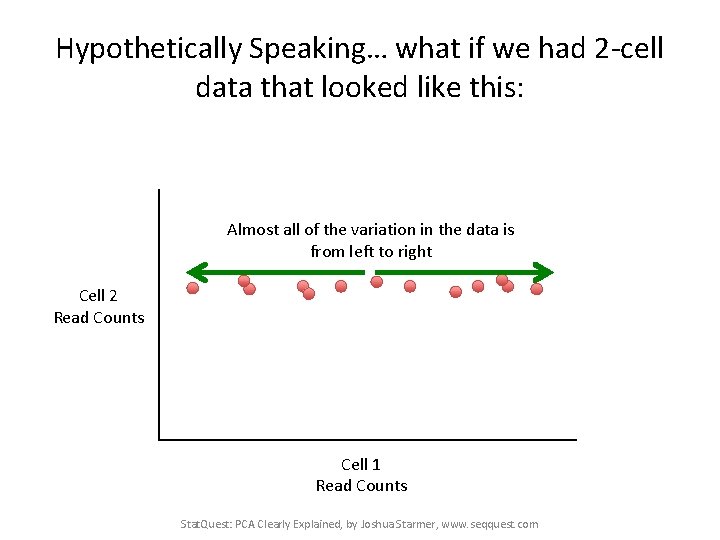
Hypothetically Speaking… what if we had 2 -cell data that looked like this: Almost all of the variation in the data is from left to right Cell 2 Read Counts Cell 1 Read Counts Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
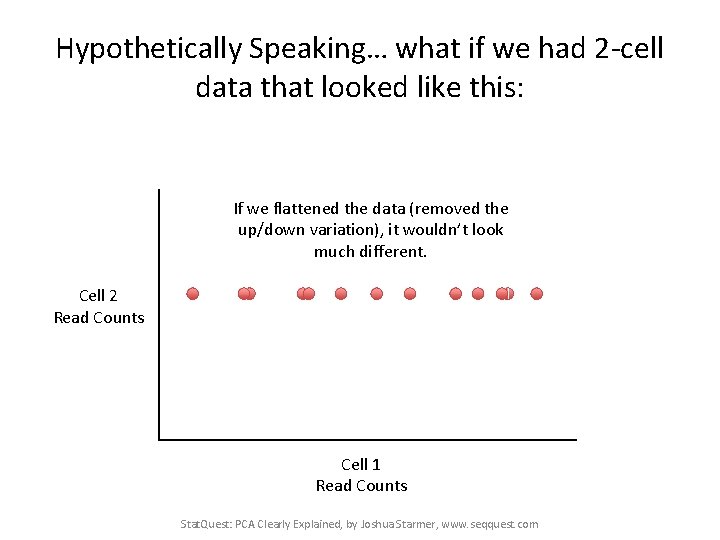
Hypothetically Speaking… what if we had 2 -cell data that looked like this: If we flattened the data (removed the up/down variation), it wouldn’t look much different. Cell 2 Read Counts Cell 1 Read Counts Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
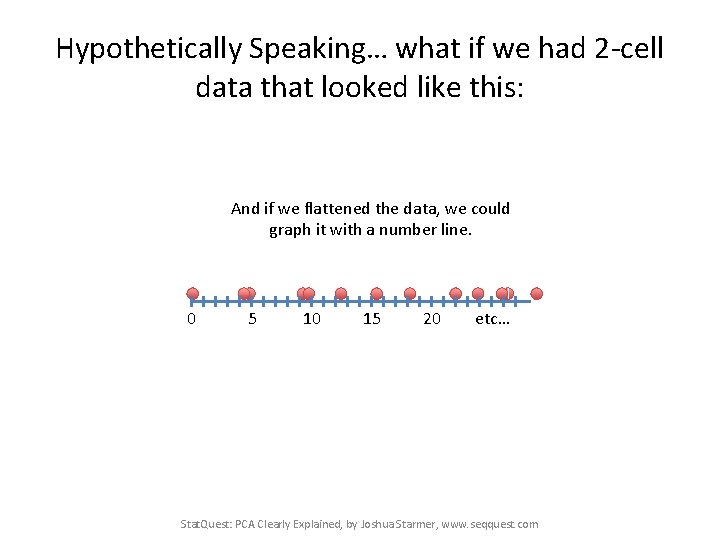
Hypothetically Speaking… what if we had 2 -cell data that looked like this: And if we flattened the data, we could graph it with a number line. 0 5 10 15 20 etc… Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
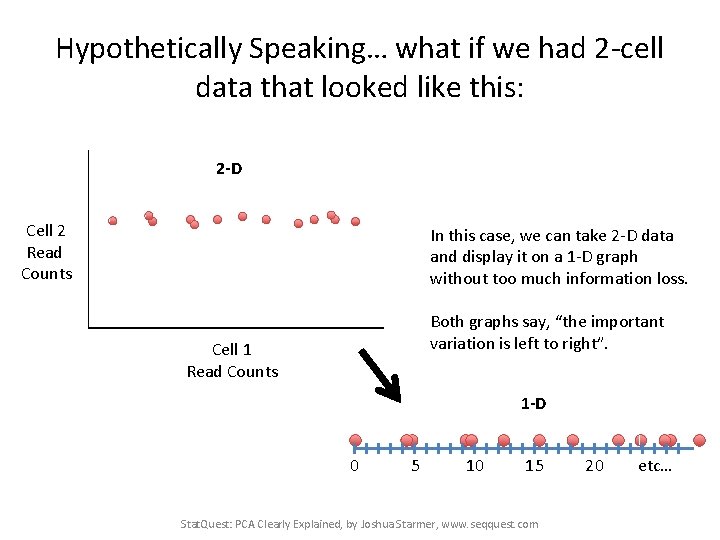
Hypothetically Speaking… what if we had 2 -cell data that looked like this: 2 -D Cell 2 Read Counts In this case, we can take 2 -D data and display it on a 1 -D graph without too much information loss. Both graphs say, “the important variation is left to right”. Cell 1 Read Counts 1 -D 0 5 10 15 Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com 20 etc…

One more example: TV and Movies are almost always 2 -D, even though the subjects are 3 -D. This is OK. The 3 rd dimension doesn’t usually add much to the story. Things still look believable without it. People look like people, things look like things, even when they have no depth and are flat on a screen. A movie camera takes 3 -D information and flattens it to 2 -D without too much loss of information. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

Summary of Dimensions • Each cell we sequence adds another “dimension” • Some dimensions are more important than others… Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
What does all of this have to do with PCA? • PCA takes a dataset with a lot of dimensions (i. e. lots of cells) and flattens it to 2 or 3 dimensions so we can look at it. – It tries to find a meaningful way to flatten the data by focusing on the things that are different between cells. (much, much more on this later) • This is sort of like flattening a Z-stack of microscope images to make a single 2 -D image for publication. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
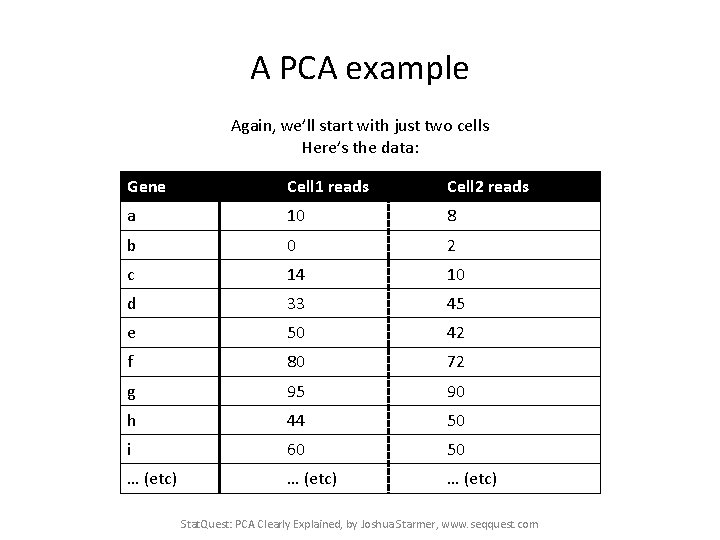
A PCA example Again, we’ll start with just two cells Here’s the data: Gene Cell 1 reads Cell 2 reads a 10 8 b 0 2 c 14 10 d 33 45 e 50 42 f 80 72 g 95 90 h 44 50 i 60 50 … (etc) Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
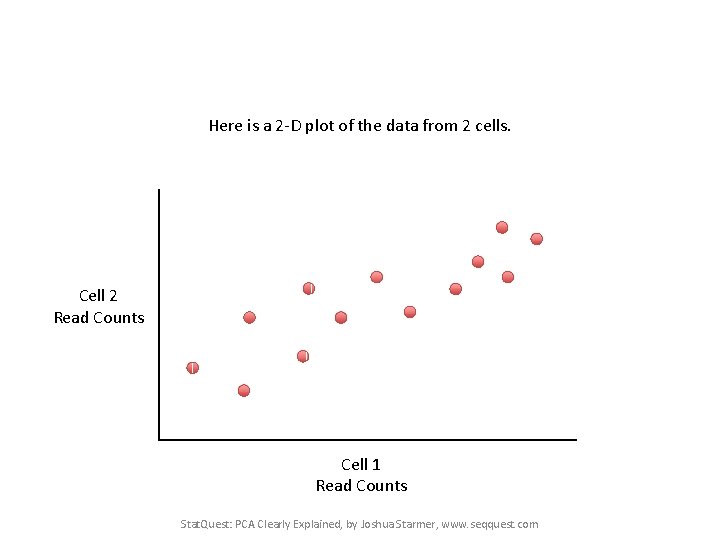
Here is a 2 -D plot of the data from 2 cells. Cell 2 Read Counts Cell 1 Read Counts Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
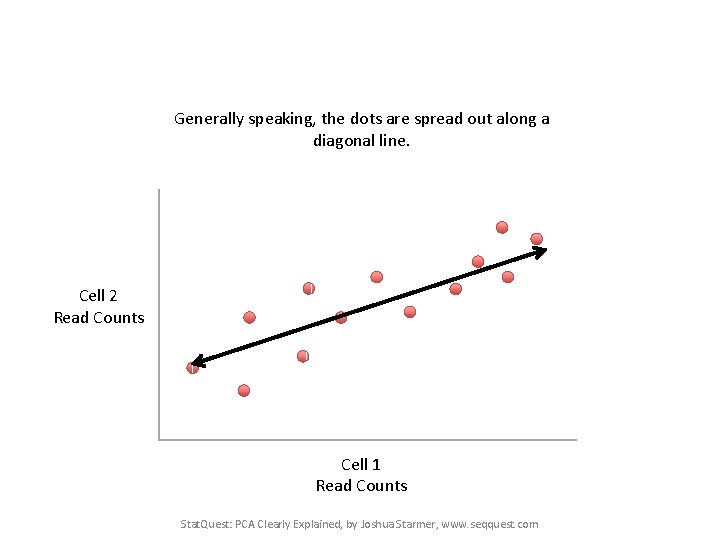
Generally speaking, the dots are spread out along a diagonal line. Cell 2 Read Counts Cell 1 Read Counts Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
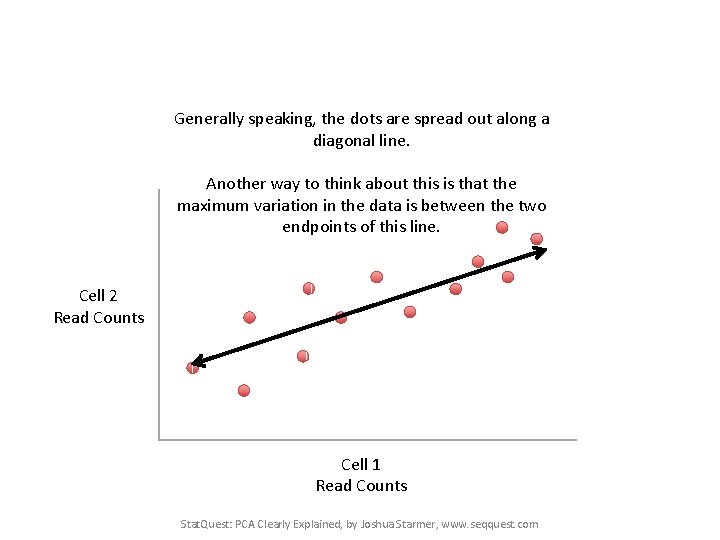
Generally speaking, the dots are spread out along a diagonal line. Another way to think about this is that the maximum variation in the data is between the two endpoints of this line. Cell 2 Read Counts Cell 1 Read Counts Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
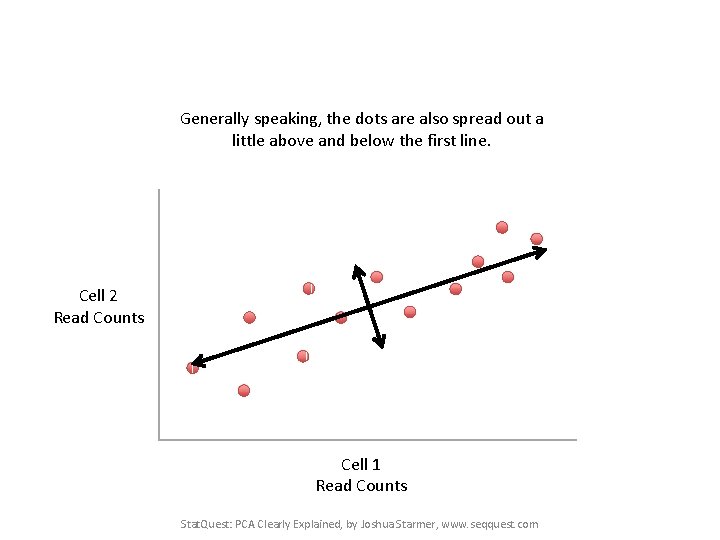
Generally speaking, the dots are also spread out a little above and below the first line. Cell 2 Read Counts Cell 1 Read Counts Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
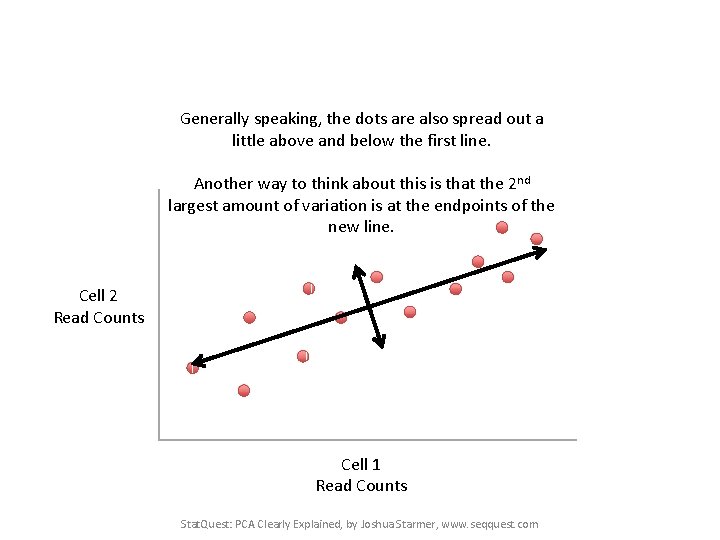
Generally speaking, the dots are also spread out a little above and below the first line. Another way to think about this is that the 2 nd largest amount of variation is at the endpoints of the new line. Cell 2 Read Counts Cell 1 Read Counts Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
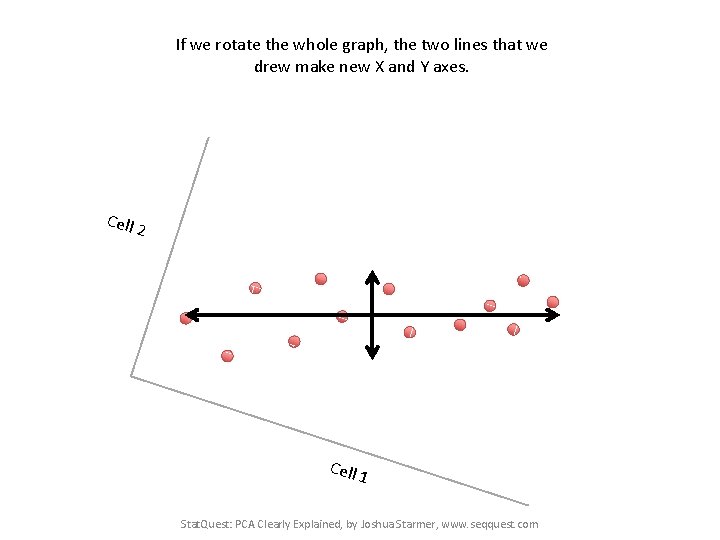
If we rotate the whole graph, the two lines that we drew make new X and Y axes. Cell 2 Cell 1 Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
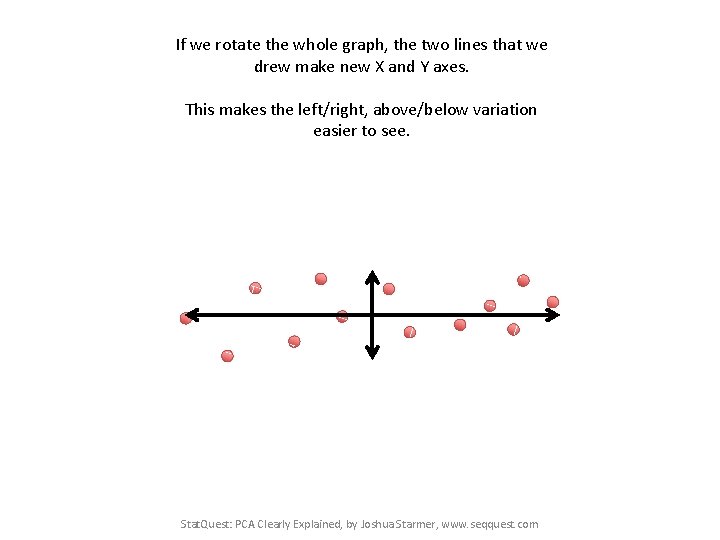
If we rotate the whole graph, the two lines that we drew make new X and Y axes. This makes the left/right, above/below variation easier to see. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
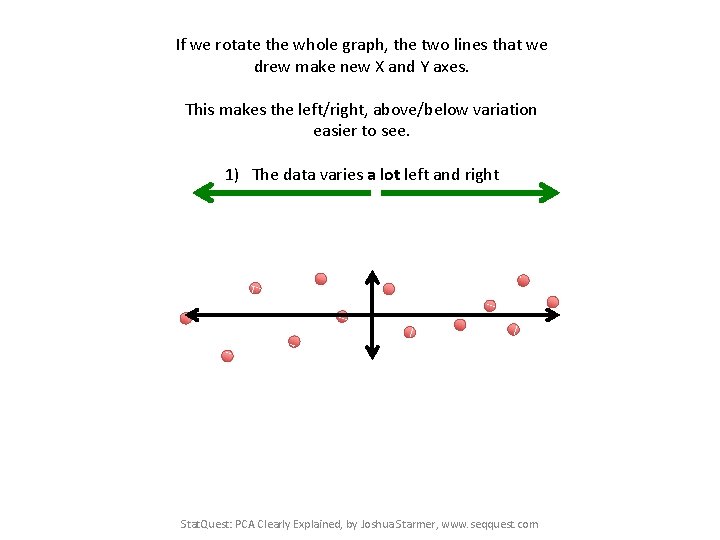
If we rotate the whole graph, the two lines that we drew make new X and Y axes. This makes the left/right, above/below variation easier to see. 1) The data varies a lot left and right Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
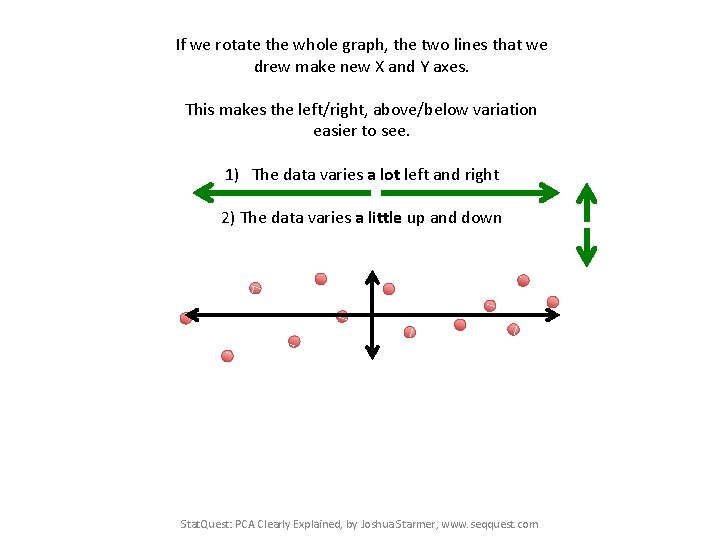
If we rotate the whole graph, the two lines that we drew make new X and Y axes. This makes the left/right, above/below variation easier to see. 1) The data varies a lot left and right 2) The data varies a little up and down Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
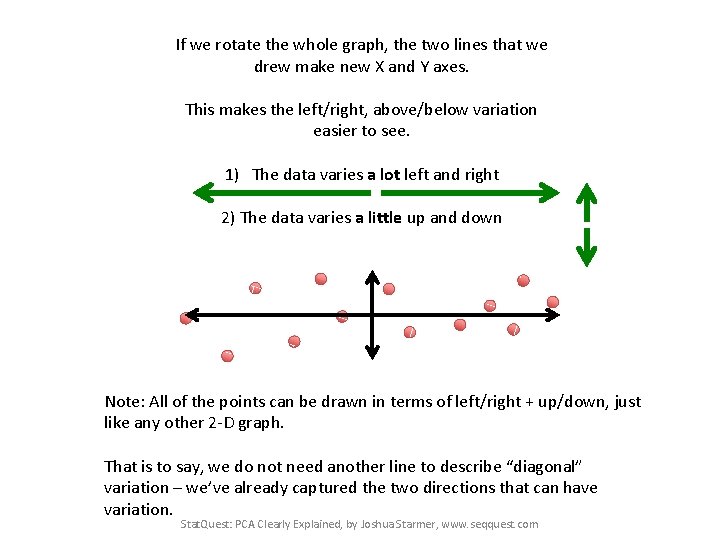
If we rotate the whole graph, the two lines that we drew make new X and Y axes. This makes the left/right, above/below variation easier to see. 1) The data varies a lot left and right 2) The data varies a little up and down Note: All of the points can be drawn in terms of left/right + up/down, just like any other 2 -D graph. That is to say, we do not need another line to describe “diagonal” variation – we’ve already captured the two directions that can have variation. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
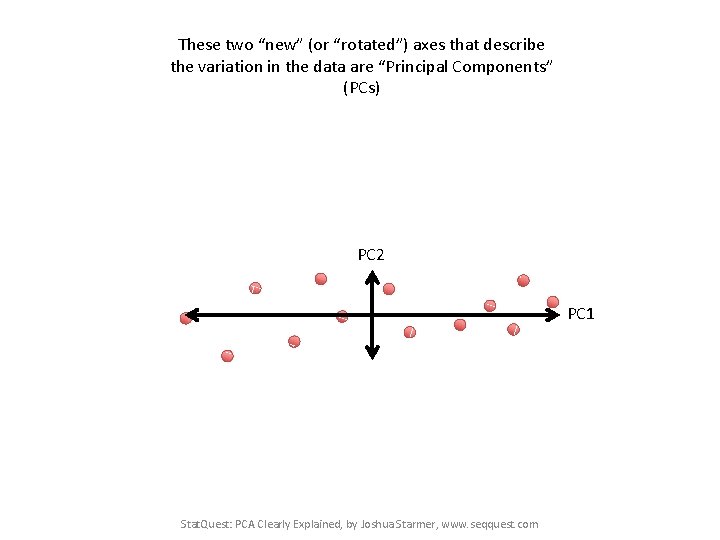
These two “new” (or “rotated”) axes that describe the variation in the data are “Principal Components” (PCs) PC 2 PC 1 Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
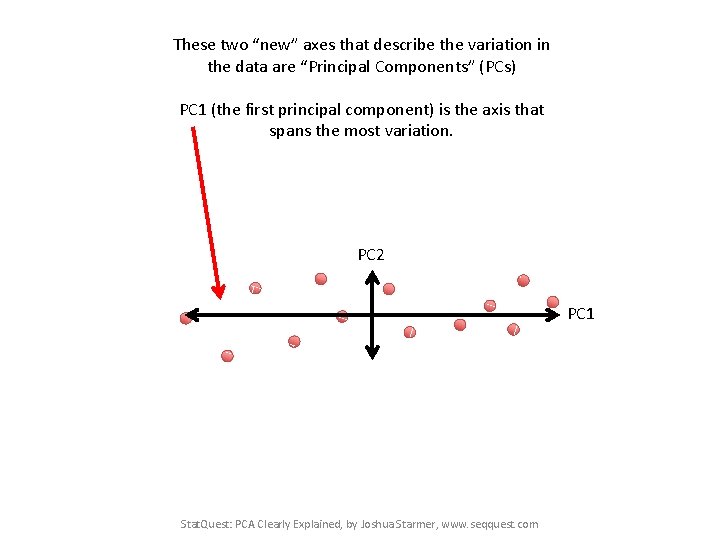
These two “new” axes that describe the variation in the data are “Principal Components” (PCs) PC 1 (the first principal component) is the axis that spans the most variation. PC 2 PC 1 Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
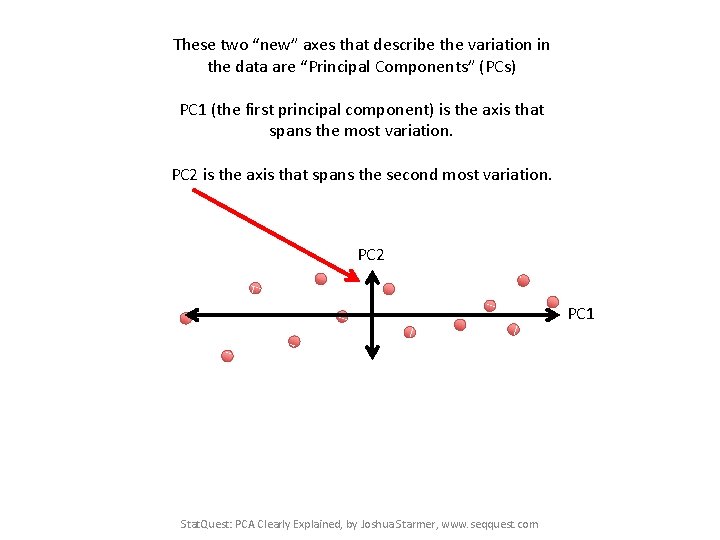
These two “new” axes that describe the variation in the data are “Principal Components” (PCs) PC 1 (the first principal component) is the axis that spans the most variation. PC 2 is the axis that spans the second most variation. PC 2 PC 1 Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

General ideas so far… Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
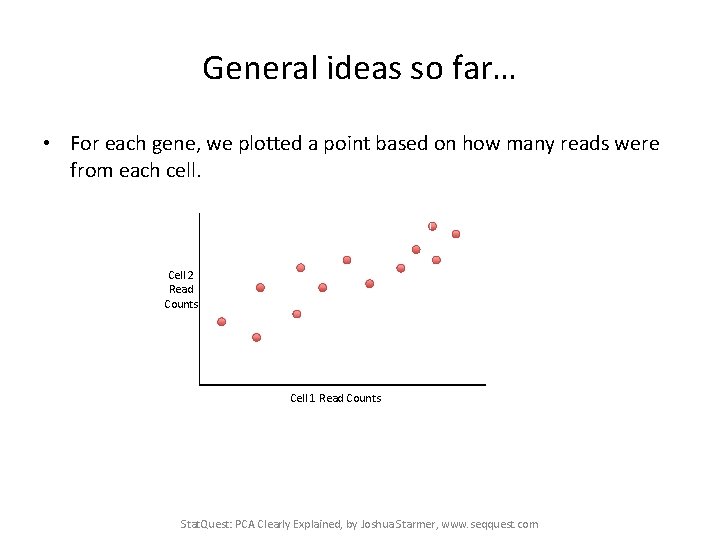
General ideas so far… • For each gene, we plotted a point based on how many reads were from each cell. Cell 2 Read Counts Cell 1 Read Counts Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
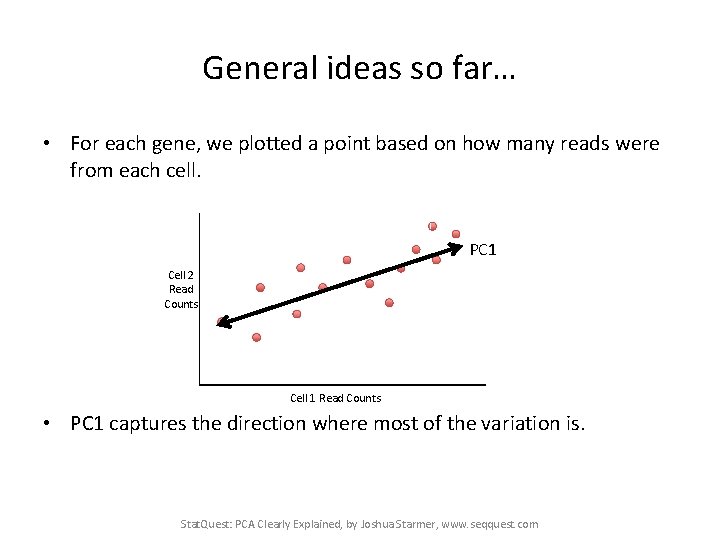
General ideas so far… • For each gene, we plotted a point based on how many reads were from each cell. PC 1 Cell 2 Read Counts Cell 1 Read Counts • PC 1 captures the direction where most of the variation is. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
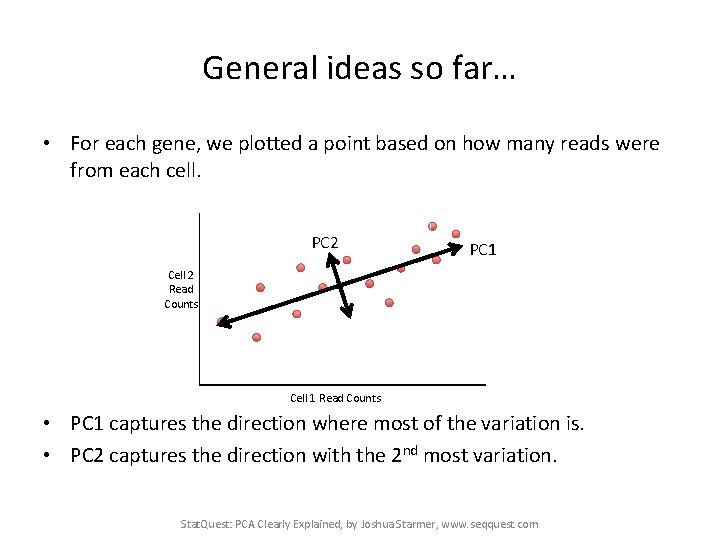
General ideas so far… • For each gene, we plotted a point based on how many reads were from each cell. PC 2 PC 1 Cell 2 Read Counts Cell 1 Read Counts • PC 1 captures the direction where most of the variation is. • PC 2 captures the direction with the 2 nd most variation. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
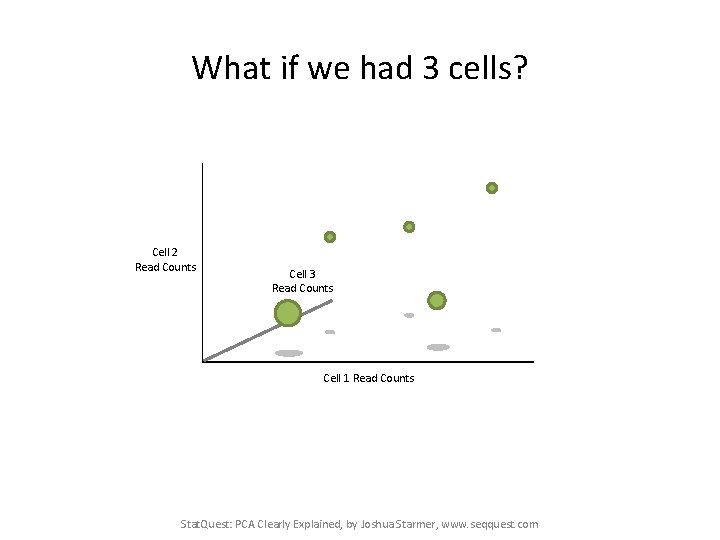
What if we had 3 cells? Cell 2 Read Counts Cell 3 Read Counts Cell 1 Read Counts Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
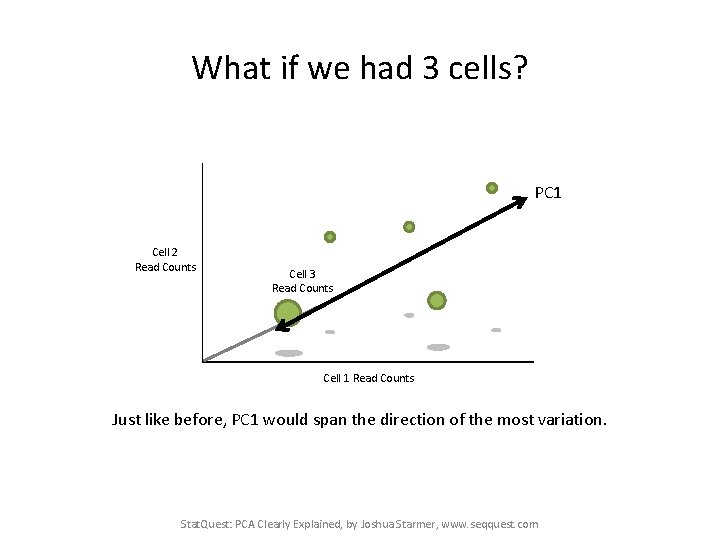
What if we had 3 cells? PC 1 Cell 2 Read Counts Cell 3 Read Counts Cell 1 Read Counts Just like before, PC 1 would span the direction of the most variation. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
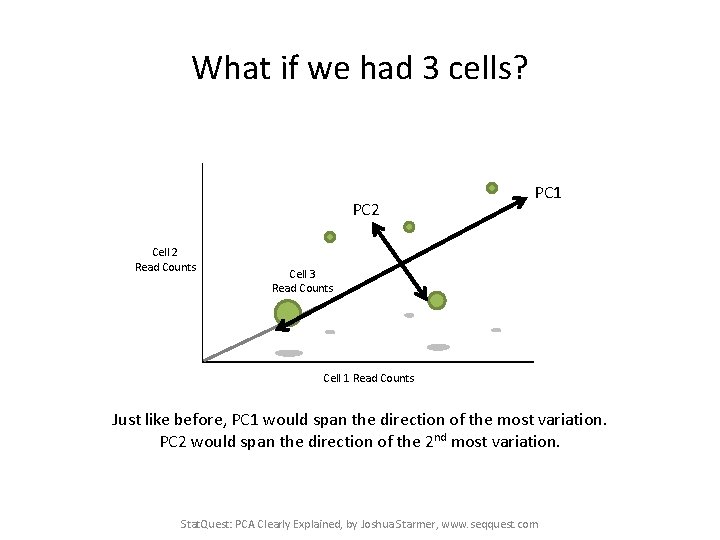
What if we had 3 cells? PC 2 Cell 2 Read Counts PC 1 Cell 3 Read Counts Cell 1 Read Counts Just like before, PC 1 would span the direction of the most variation. PC 2 would span the direction of the 2 nd most variation. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
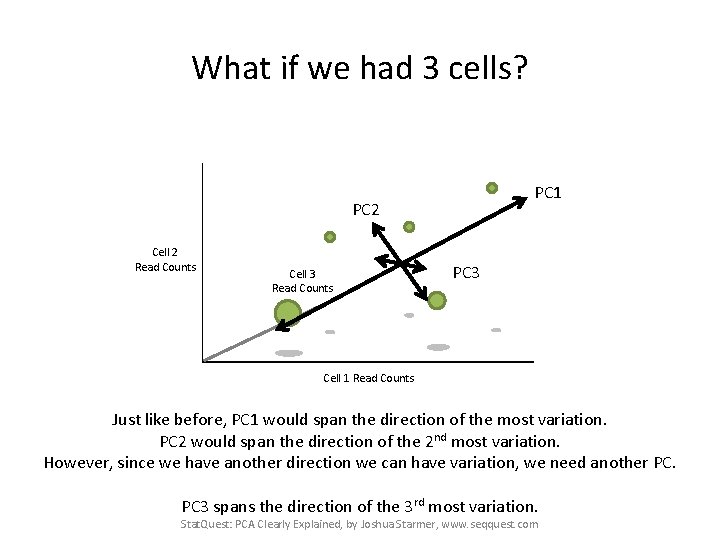
What if we had 3 cells? PC 1 PC 2 Cell 2 Read Counts Cell 3 Read Counts PC 3 Cell 1 Read Counts Just like before, PC 1 would span the direction of the most variation. PC 2 would span the direction of the 2 nd most variation. However, since we have another direction we can have variation, we need another PC. PC 3 spans the direction of the 3 rd most variation. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

What if we had 4 cells? Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

What if we had 4 cells? • PC 1 would span the direction of the most variation. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

What if we had 4 cells? • PC 1 would span the direction of the most variation. • PC 2 would span the direction of the 2 nd most variation. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

What if we had 4 cells? • PC 1 would span the direction of the most variation. • PC 2 would span the direction of the 2 nd most variation. • PC 3 would span the direction of the 3 rd most variation. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

What if we had 4 cells? • • PC 1 would span the direction of the most variation. PC 2 would span the direction of the 2 nd most variation. PC 3 would span the direction of the 3 rd most variation. PC 4 would span the direction of the 4 th most variation. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

What if we had 4 cells? • • PC 1 would span the direction of the most variation. PC 2 would span the direction of the 2 nd most variation. PC 3 would span the direction of the 3 rd most variation. PC 4 would span the direction of the 4 th most variation. There is a principal component for each dimension (cell). If we had 200 cells, we would have 200 principal components. PC 200 would span the direction of the 200 th most variation. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
Examples of PCs PC 2 PC 1 Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
Examples of PCs PC 2 PC 1 PC 2 Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
Examples of PCs PC 1 PC 2 PC 1 Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
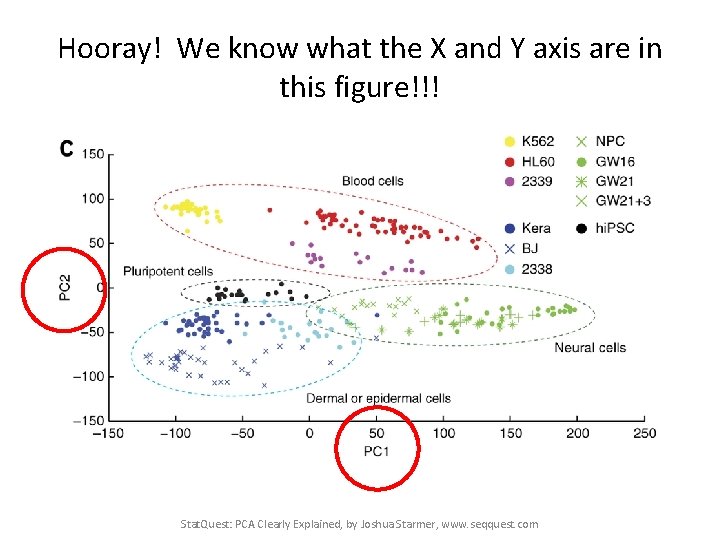
Hooray! We know what the X and Y axis are in this figure!!! Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
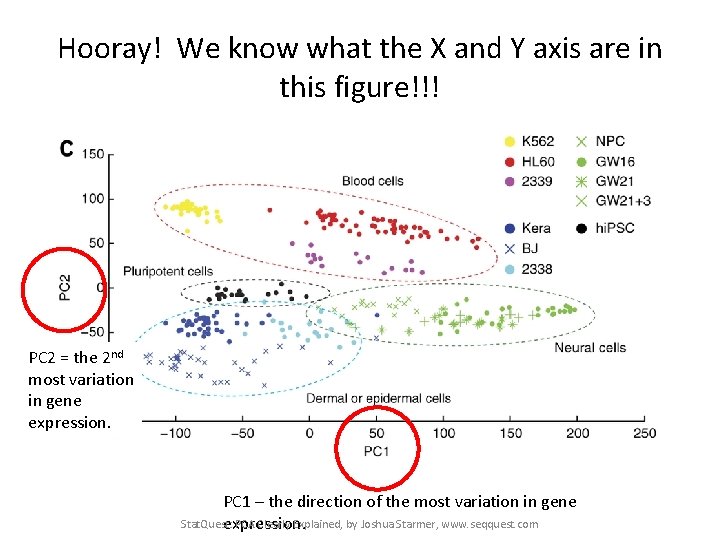
Hooray! We know what the X and Y axis are in this figure!!! PC 2 = the 2 nd most variation in gene expression. PC 1 – the direction of the most variation in gene Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com expression.
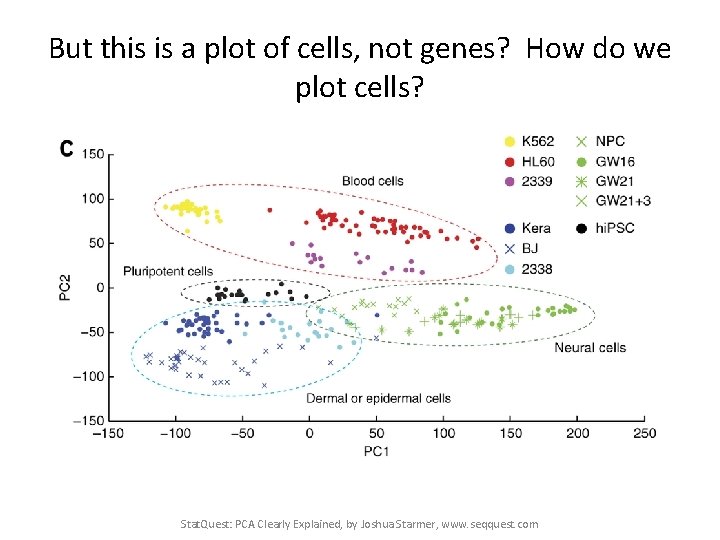
But this is a plot of cells, not genes? How do we plot cells? Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
Back to the original scatter plot… PC 1 PC 2 Cell 1 Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
For now, let’s focus on PC 1 Cell 2 Cell 1 Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
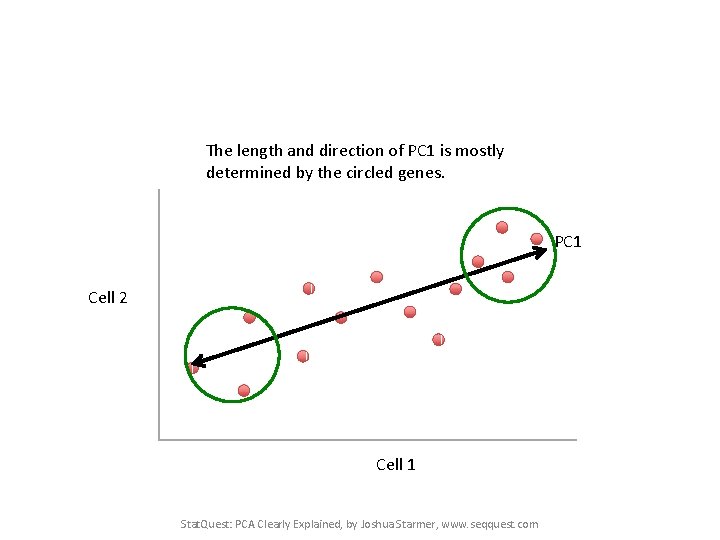
The length and direction of PC 1 is mostly determined by the circled genes. PC 1 Cell 2 Cell 1 Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
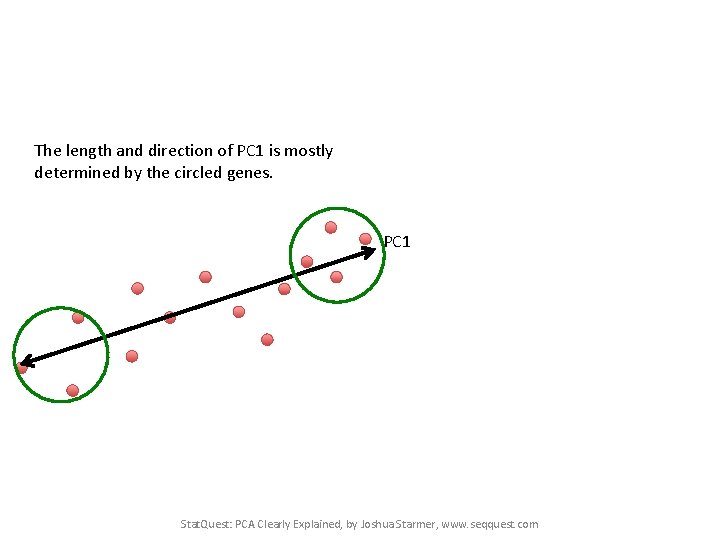
The length and direction of PC 1 is mostly determined by the circled genes. PC 1 Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
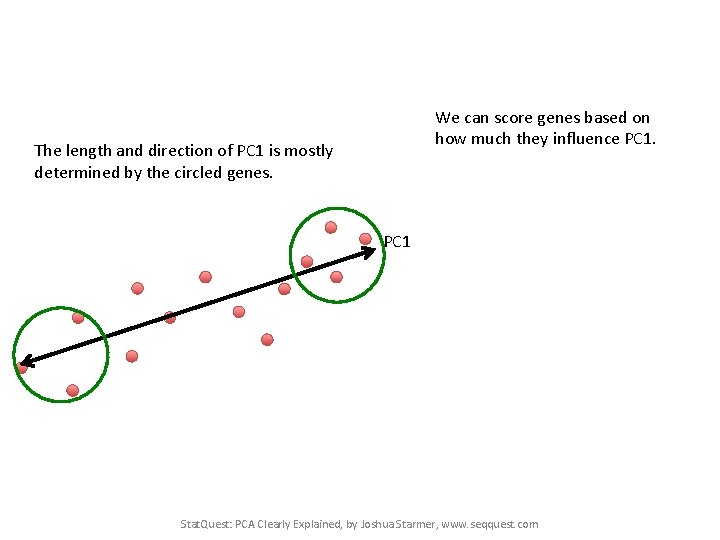
We can score genes based on how much they influence PC 1. The length and direction of PC 1 is mostly determined by the circled genes. PC 1 Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
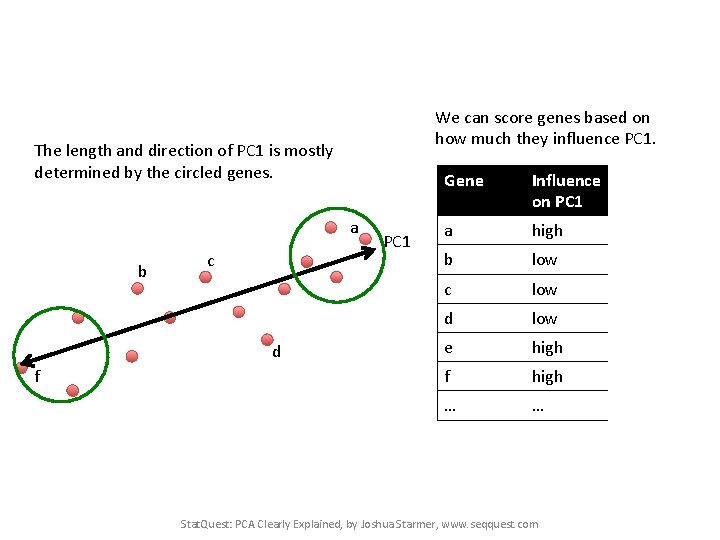
We can score genes based on how much they influence PC 1. The length and direction of PC 1 is mostly determined by the circled genes. a b c d f PC 1 Gene Influence In on PC 1 numbers a high 10 b low 0. 5 c low 3 d low -0. 2 e high 13 f high -14 … … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
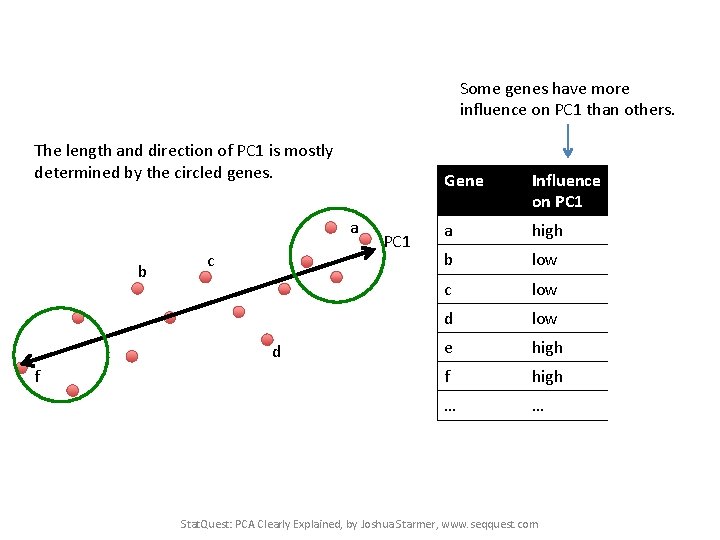
Some genes have more influence on PC 1 than others. The length and direction of PC 1 is mostly determined by the circled genes. a b c d f PC 1 Gene Influence In on PC 1 numbers a high 10 b low 0. 5 c low 3 d low -0. 2 e high 13 f high -14 … … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
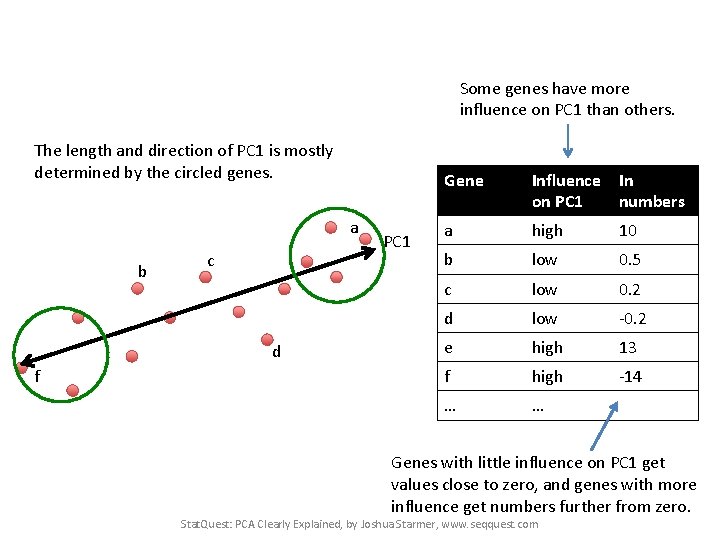
Some genes have more influence on PC 1 than others. The length and direction of PC 1 is mostly determined by the circled genes. a b c d f PC 1 Gene Influence In on PC 1 numbers a high 10 b low 0. 5 c low 0. 2 d low -0. 2 e high 13 f high -14 … … Genes with little influence on PC 1 get values close to zero, and genes with more influence get numbers further from zero. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
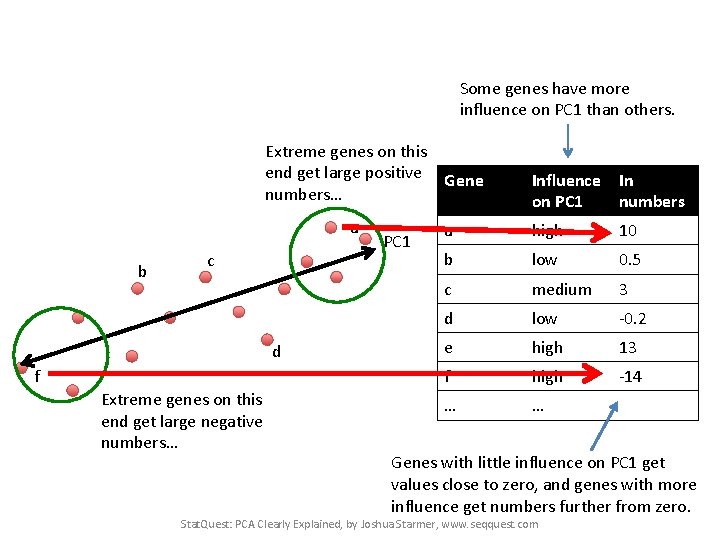
Some genes have more influence on PC 1 than others. Extreme genes on this end get large positive Gene numbers… a b c d f Extreme genes on this end get large negative numbers… PC 1 Influence In on PC 1 numbers a high 10 b low 0. 5 c medium 3 d low -0. 2 e high 13 f high -14 … … Genes with little influence on PC 1 get values close to zero, and genes with more influence get numbers further from zero. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
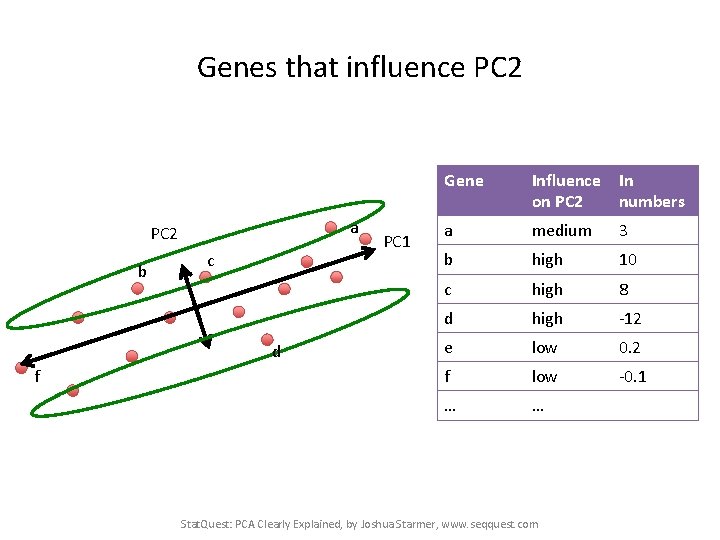
Genes that influence PC 2 a PC 2 b c d f PC 1 Gene Influence In on PC 2 numbers a medium 3 b high 10 c high 8 d high -12 e low 0. 2 f low -0. 1 … … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
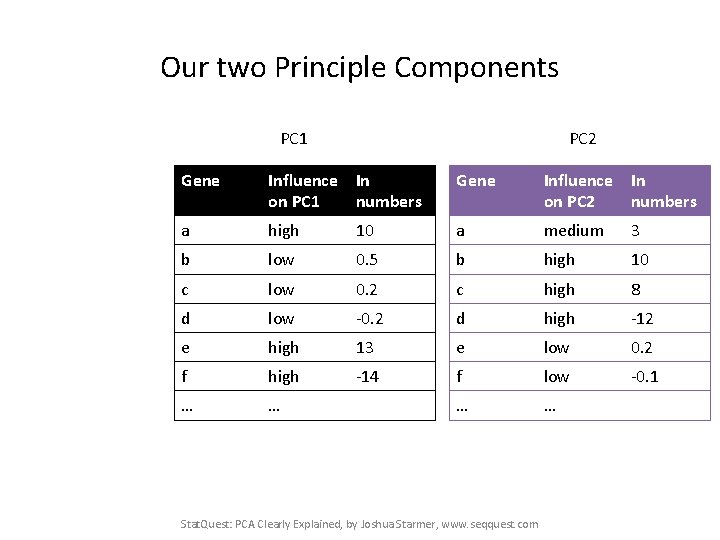
Our two Principle Components PC 1 PC 2 Gene Influence In on PC 1 numbers Gene Influence In on PC 2 numbers a high 10 a medium 3 b low 0. 5 b high 10 c low 0. 2 c high 8 d low -0. 2 d high -12 e high 13 e low 0. 2 f high -14 f low -0. 1 … … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
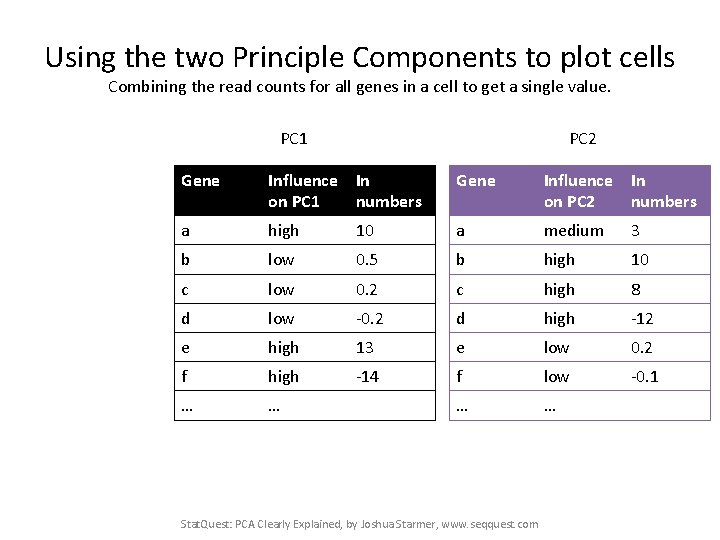
Using the two Principle Components to plot cells Combining the read counts for all genes in a cell to get a single value. PC 1 PC 2 Gene Influence In on PC 1 numbers Gene Influence In on PC 2 numbers a high 10 a medium 3 b low 0. 5 b high 10 c low 0. 2 c high 8 d low -0. 2 d high -12 e high 13 e low 0. 2 f high -14 f low -0. 1 … … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
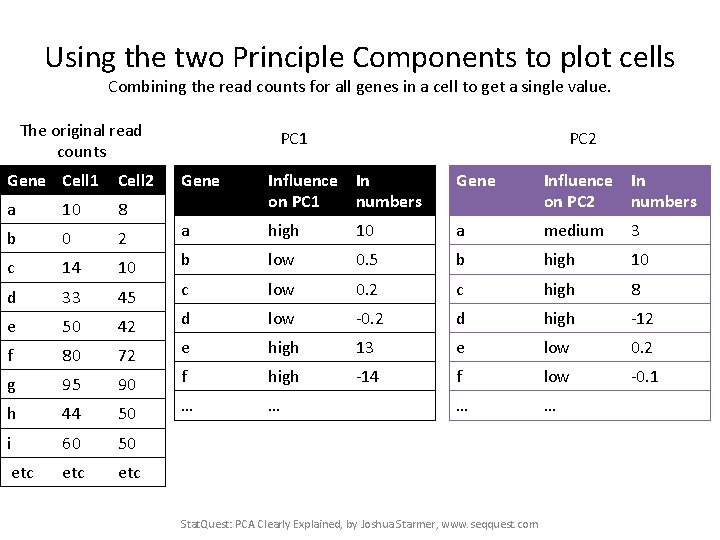
Using the two Principle Components to plot cells Combining the read counts for all genes in a cell to get a single value. The original read counts Gene Cell 1 Cell 2 a 10 8 b 0 2 c 14 10 d 33 45 e 50 42 f 80 72 g 95 90 h 44 50 i 60 50 etc etc PC 1 PC 2 Gene Influence In on PC 1 numbers Gene Influence In on PC 2 numbers a high 10 a medium 3 b low 0. 5 b high 10 c low 0. 2 c high 8 d low -0. 2 d high -12 e high 13 e low 0. 2 f high -14 f low -0. 1 … … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
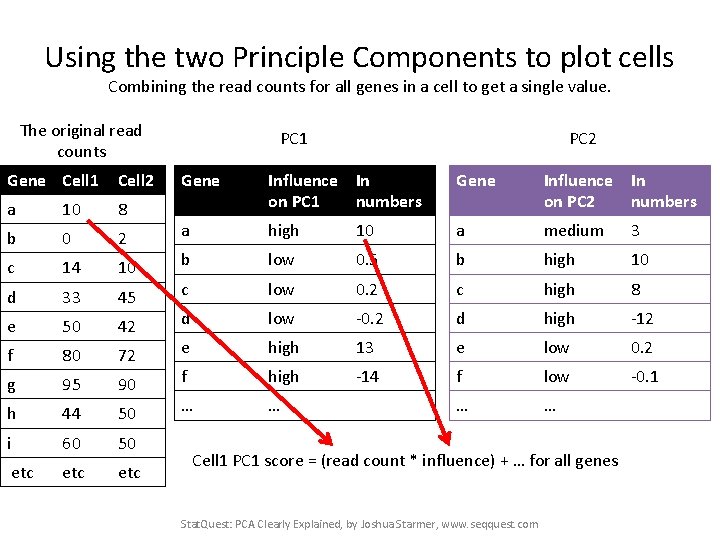
Using the two Principle Components to plot cells Combining the read counts for all genes in a cell to get a single value. The original read counts Gene Cell 1 Cell 2 a 10 8 b 0 2 c 14 10 d 33 45 e 50 42 f 80 72 g 95 90 h 44 50 i 60 50 etc etc PC 1 PC 2 Gene Influence In on PC 1 numbers Gene Influence In on PC 2 numbers a high 10 a medium 3 b low 0. 5 b high 10 c low 0. 2 c high 8 d low -0. 2 d high -12 e high 13 e low 0. 2 f high -14 f low -0. 1 … … Cell 1 PC 1 score = (read count * influence) + … for all genes Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
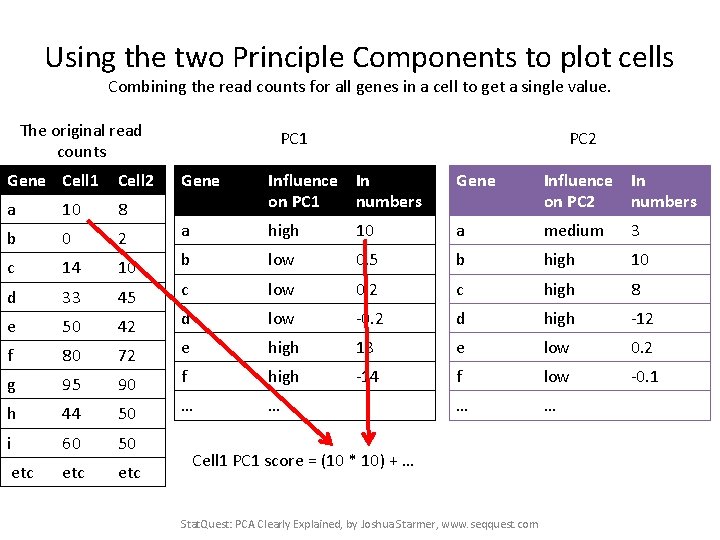
Using the two Principle Components to plot cells Combining the read counts for all genes in a cell to get a single value. The original read counts Gene Cell 1 Cell 2 a 10 8 b 0 2 c 14 10 d 33 45 e 50 42 f 80 72 g 95 90 h 44 50 i 60 50 etc etc PC 1 PC 2 Gene Influence In on PC 1 numbers Gene Influence In on PC 2 numbers a high 10 a medium 3 b low 0. 5 b high 10 c low 0. 2 c high 8 d low -0. 2 d high -12 e high 13 e low 0. 2 f high -14 f low -0. 1 … … Cell 1 PC 1 score = (10 * 10) + … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
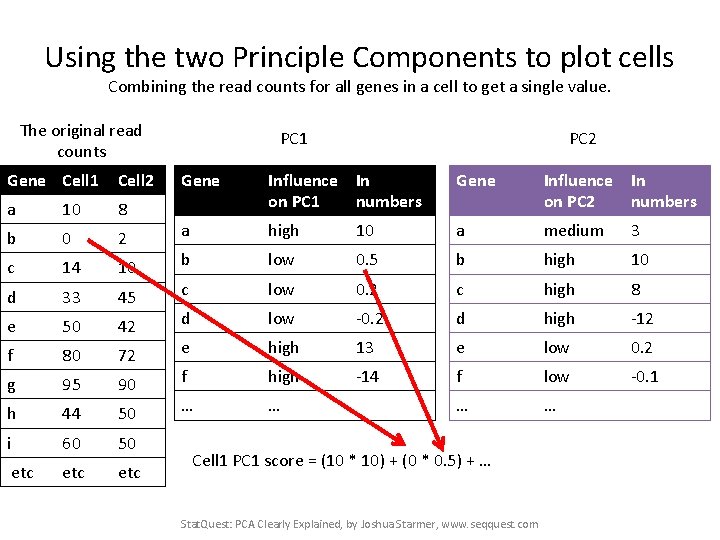
Using the two Principle Components to plot cells Combining the read counts for all genes in a cell to get a single value. The original read counts Gene Cell 1 Cell 2 a 10 8 b 0 2 c 14 10 d 33 45 e 50 42 f 80 72 g 95 90 h 44 50 i 60 50 etc etc PC 1 PC 2 Gene Influence In on PC 1 numbers Gene Influence In on PC 2 numbers a high 10 a medium 3 b low 0. 5 b high 10 c low 0. 2 c high 8 d low -0. 2 d high -12 e high 13 e low 0. 2 f high -14 f low -0. 1 … … Cell 1 PC 1 score = (10 * 10) + (0 * 0. 5) + … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
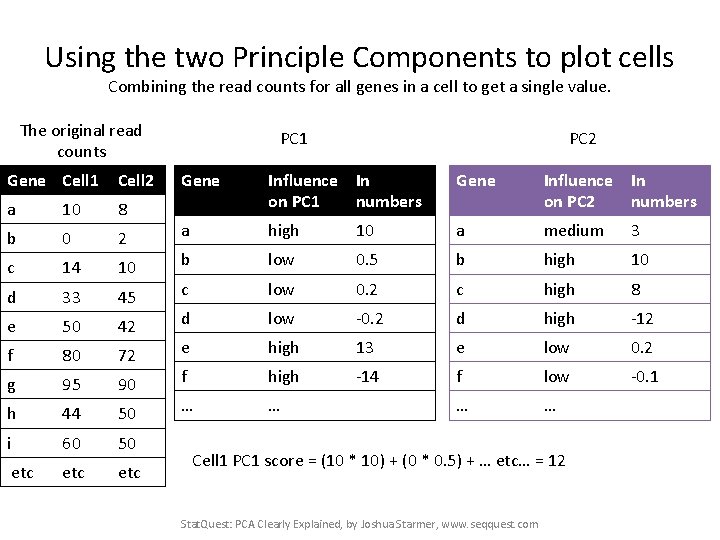
Using the two Principle Components to plot cells Combining the read counts for all genes in a cell to get a single value. The original read counts Gene Cell 1 Cell 2 a 10 8 b 0 2 c 14 10 d 33 45 e 50 42 f 80 72 g 95 90 h 44 50 i 60 50 etc etc PC 1 PC 2 Gene Influence In on PC 1 numbers Gene Influence In on PC 2 numbers a high 10 a medium 3 b low 0. 5 b high 10 c low 0. 2 c high 8 d low -0. 2 d high -12 e high 13 e low 0. 2 f high -14 f low -0. 1 … … Cell 1 PC 1 score = (10 * 10) + (0 * 0. 5) + … etc… = 12 Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
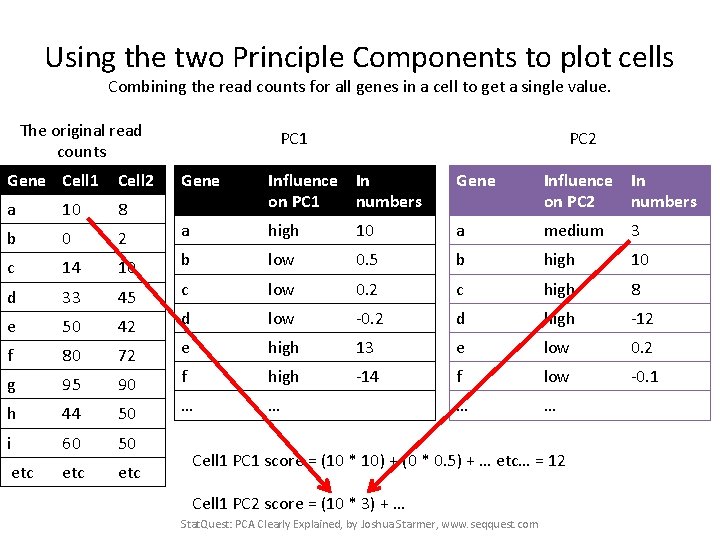
Using the two Principle Components to plot cells Combining the read counts for all genes in a cell to get a single value. The original read counts Gene Cell 1 Cell 2 a 10 8 b 0 2 c 14 10 d 33 45 e 50 42 f 80 72 g 95 90 h 44 50 i 60 50 etc etc PC 1 PC 2 Gene Influence In on PC 1 numbers Gene Influence In on PC 2 numbers a high 10 a medium 3 b low 0. 5 b high 10 c low 0. 2 c high 8 d low -0. 2 d high -12 e high 13 e low 0. 2 f high -14 f low -0. 1 … … Cell 1 PC 1 score = (10 * 10) + (0 * 0. 5) + … etc… = 12 Cell 1 PC 2 score = (10 * 3) + … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
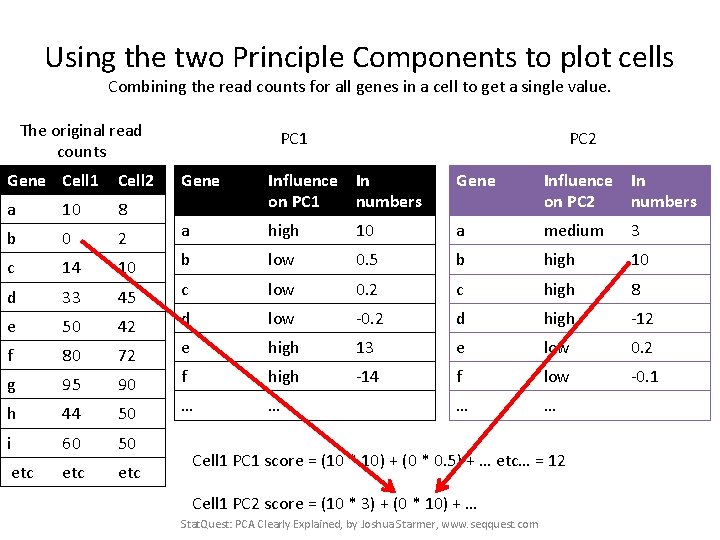
Using the two Principle Components to plot cells Combining the read counts for all genes in a cell to get a single value. The original read counts Gene Cell 1 Cell 2 a 10 8 b 0 2 c 14 10 d 33 45 e 50 42 f 80 72 g 95 90 h 44 50 i 60 50 etc etc PC 1 PC 2 Gene Influence In on PC 1 numbers Gene Influence In on PC 2 numbers a high 10 a medium 3 b low 0. 5 b high 10 c low 0. 2 c high 8 d low -0. 2 d high -12 e high 13 e low 0. 2 f high -14 f low -0. 1 … … Cell 1 PC 1 score = (10 * 10) + (0 * 0. 5) + … etc… = 12 Cell 1 PC 2 score = (10 * 3) + (0 * 10) + … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
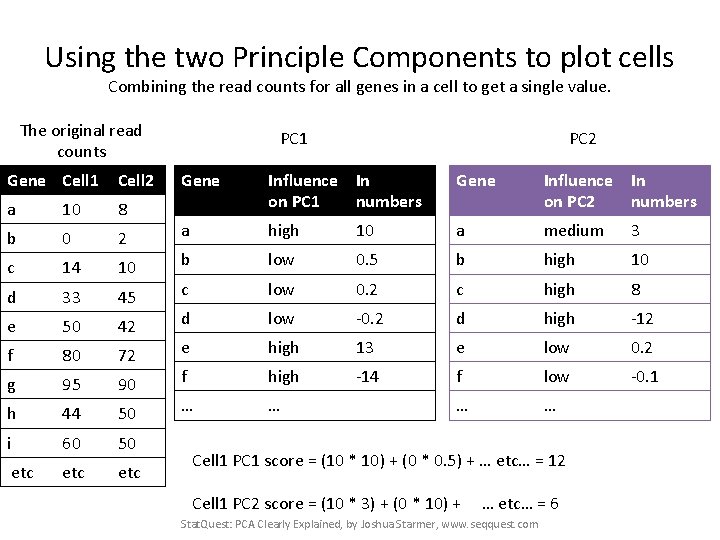
Using the two Principle Components to plot cells Combining the read counts for all genes in a cell to get a single value. The original read counts Gene Cell 1 Cell 2 a 10 8 b 0 2 c 14 10 d 33 45 e 50 42 f 80 72 g 95 90 h 44 50 i 60 50 etc etc PC 1 PC 2 Gene Influence In on PC 1 numbers Gene Influence In on PC 2 numbers a high 10 a medium 3 b low 0. 5 b high 10 c low 0. 2 c high 8 d low -0. 2 d high -12 e high 13 e low 0. 2 f high -14 f low -0. 1 … … Cell 1 PC 1 score = (10 * 10) + (0 * 0. 5) + … etc… = 12 Cell 1 PC 2 score = (10 * 3) + (0 * 10) + … etc… = 6 Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

Cell 1 PC 1 score = (10 * 10) + (0 * 0. 5) + … etc… = 12 Cell 1 PC 2 score = (10 * 3) + (0 * 10) + … etc… = 6 Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
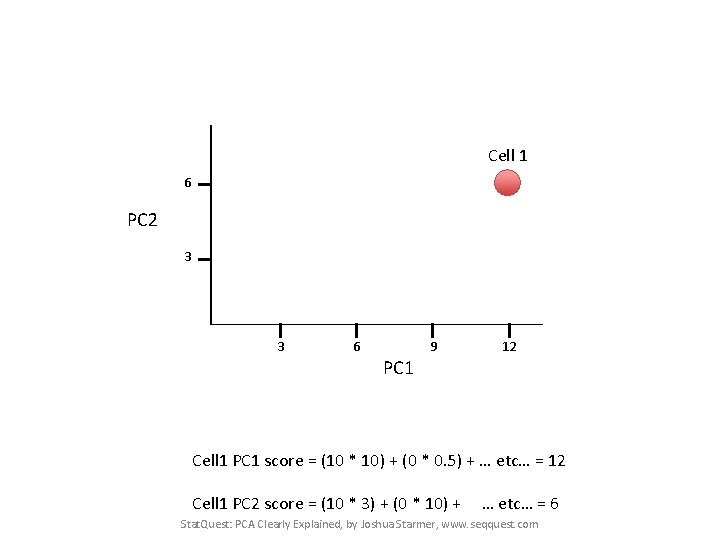
Cell 1 6 PC 2 3 3 6 9 12 PC 1 Cell 1 PC 1 score = (10 * 10) + (0 * 0. 5) + … etc… = 12 Cell 1 PC 2 score = (10 * 3) + (0 * 10) + … etc… = 6 Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
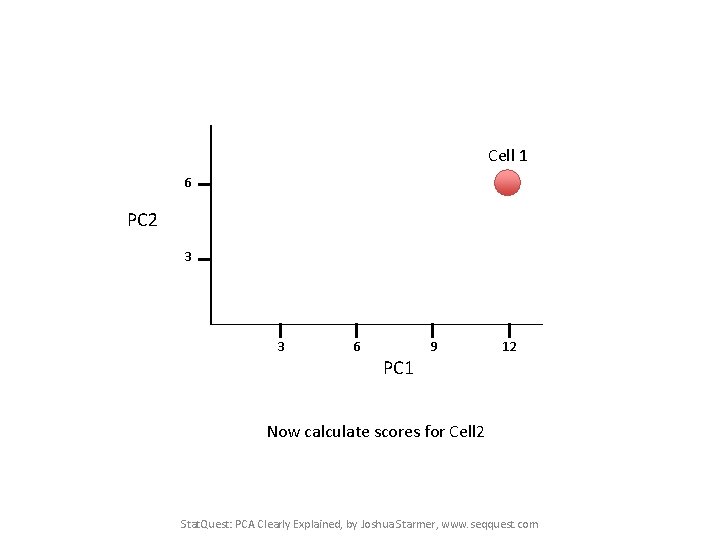
Cell 1 6 PC 2 3 3 6 9 12 PC 1 Now calculate scores for Cell 2 Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
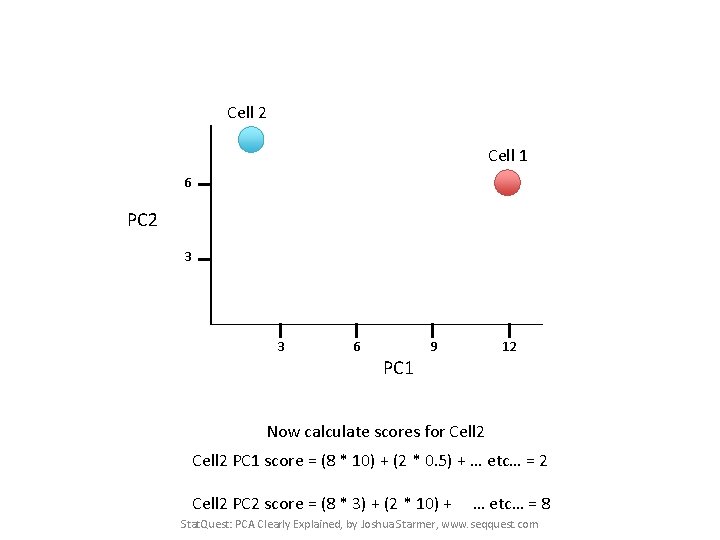
Cell 2 Cell 1 6 PC 2 3 3 6 9 12 PC 1 Now calculate scores for Cell 2 PC 1 score = (8 * 10) + (2 * 0. 5) + … etc… = 2 Cell 2 PC 2 score = (8 * 3) + (2 * 10) + … etc… = 8 Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
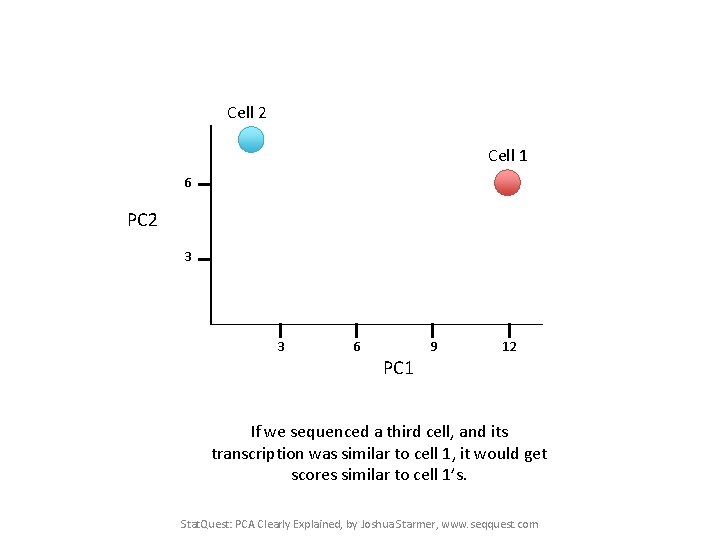
Cell 2 Cell 1 6 PC 2 3 3 6 9 12 PC 1 If we sequenced a third cell, and its transcription was similar to cell 1, it would get scores similar to cell 1’s. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
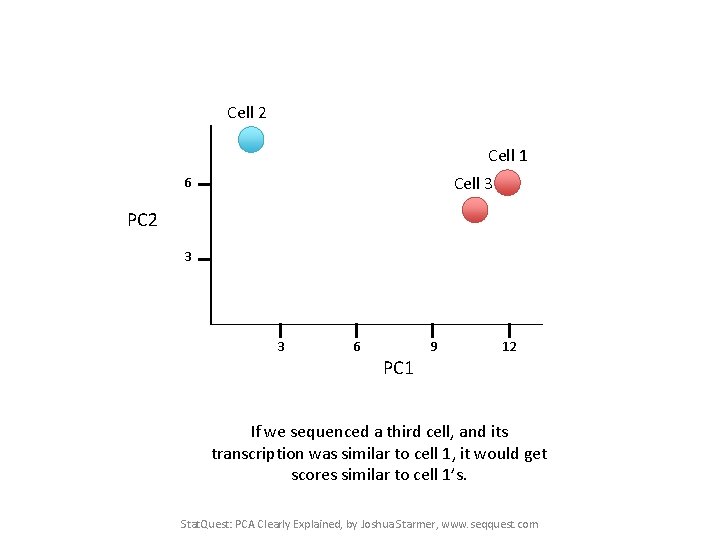
Cell 2 Cell 1 Cell 3 6 PC 2 3 3 6 9 12 PC 1 If we sequenced a third cell, and its transcription was similar to cell 1, it would get scores similar to cell 1’s. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
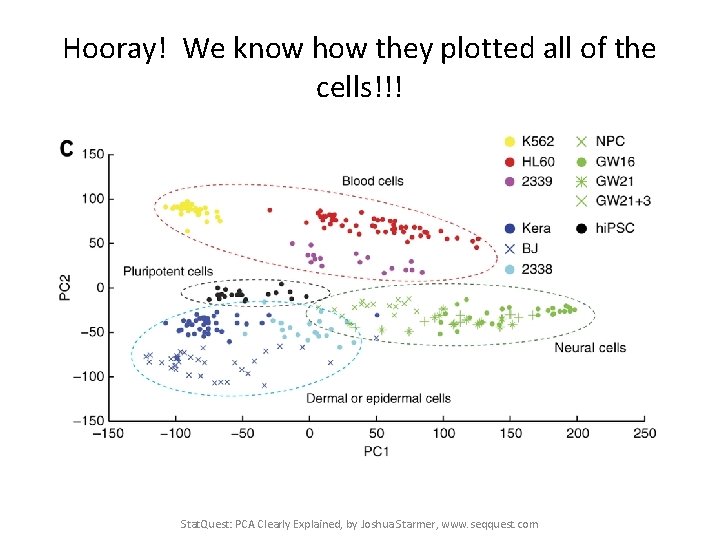
Hooray! We know how they plotted all of the cells!!! Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

General ideas so far… • Genes with the largest variation between cells will have the most influence on the principal components. – i. e. genes highly expressed in some cells and not expressed in others will have a lot of variation and influence on the PCs. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
General ideas so far… • Genes with the largest variation between cells will have the most influence on the principal components. – i. e. genes highly expressed in some cells and not expressed in others will have a lot of variation and influence on the PCs. • The 1 st PC captures the most variation in the data. • The 2 nd PC captures the second most variation in the data. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
General ideas so far… • Genes with the largest variation between cells will have the most influence on the principal components. – i. e. genes highly expressed in some cells and not expressed in others will have a lot of variation and influence on the PCs. • The 1 st PC captures the most variation in the data. • The 2 nd PC captures the second most variation in the data. • You can use the original data and the first 2 PCs to get X/Y values to plot on a figure. Cells with similar transcription patterns will cluster together. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

But wait, there’s more!!! Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
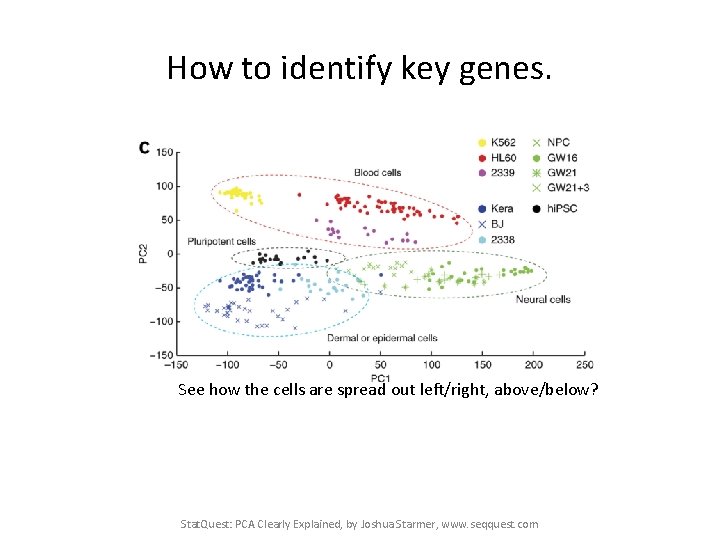
How to identify key genes. See how the cells are spread out left/right, above/below? Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
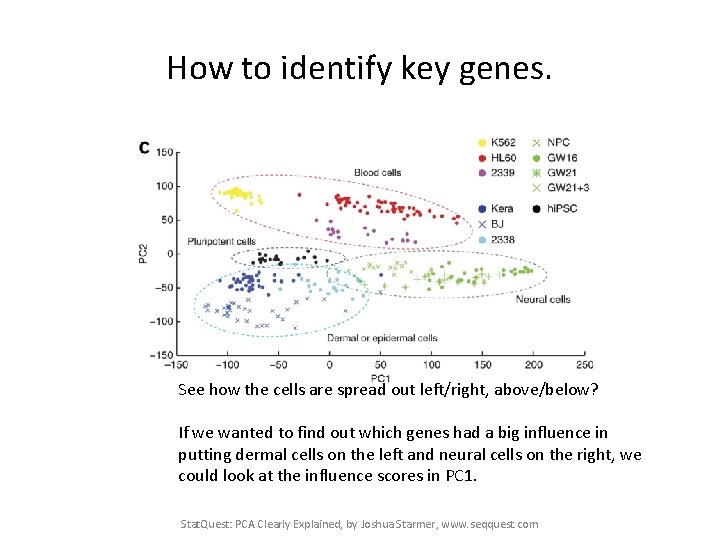
How to identify key genes. See how the cells are spread out left/right, above/below? If we wanted to find out which genes had a big influence in putting dermal cells on the left and neural cells on the right, we could look at the influence scores in PC 1. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
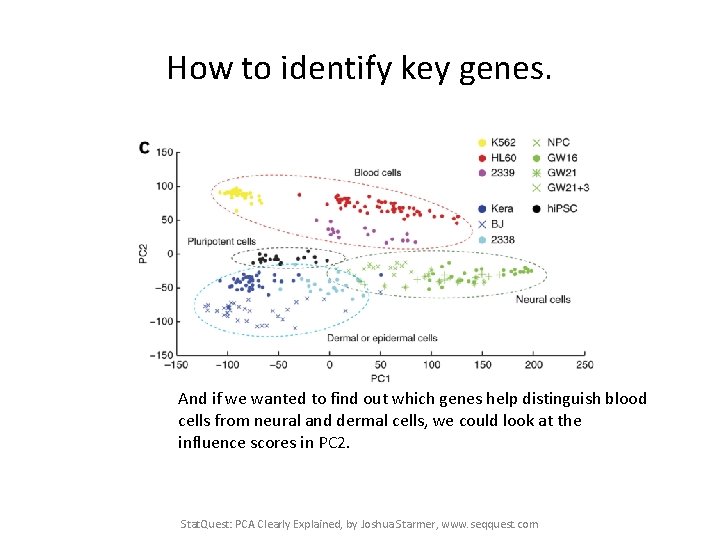
How to identify key genes. And if we wanted to find out which genes help distinguish blood cells from neural and dermal cells, we could look at the influence scores in PC 2. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com

But wait, there’s even more? Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
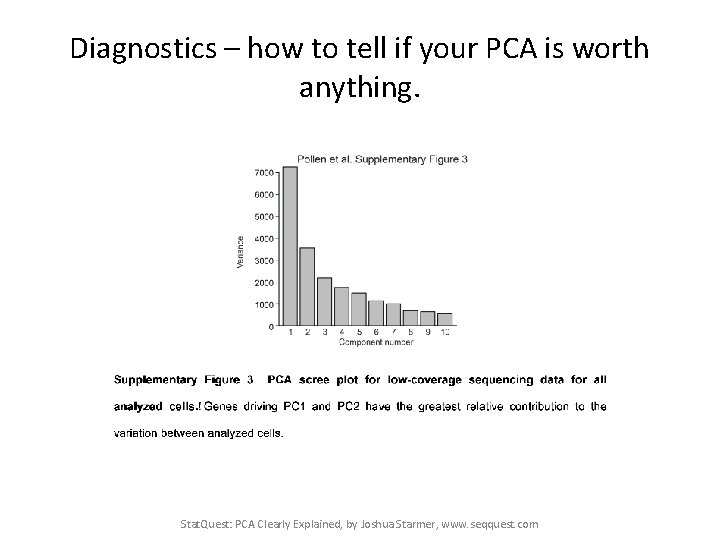
Diagnostics – how to tell if your PCA is worth anything. Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
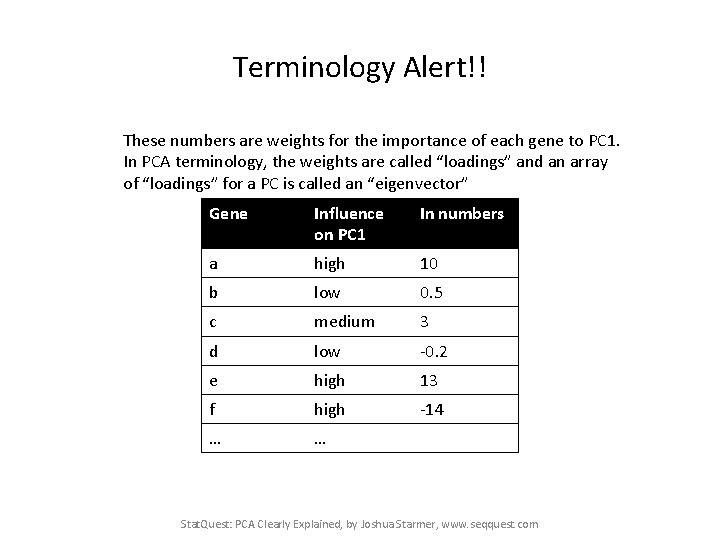
Terminology Alert!! These numbers are weights for the importance of each gene to PC 1. In PCA terminology, the weights are called “loadings” and an array of “loadings” for a PC is called an “eigenvector” Gene Influence on PC 1 In numbers a high 10 b low 0. 5 c medium 3 d low -0. 2 e high 13 f high -14 … … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com
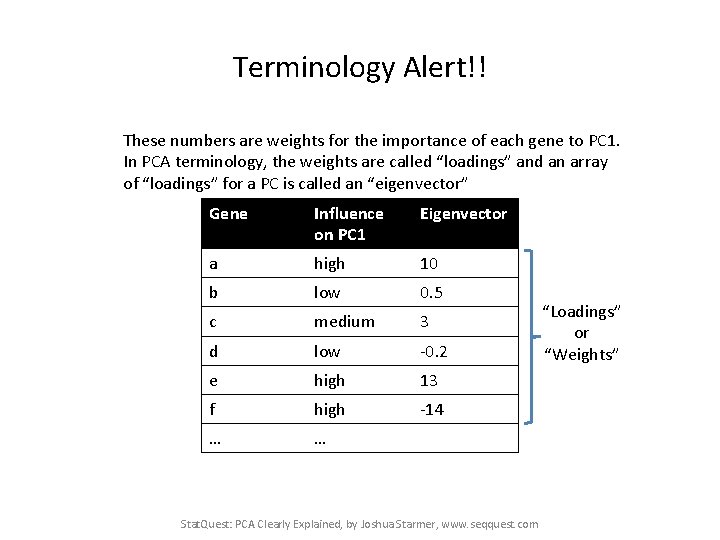
Terminology Alert!! These numbers are weights for the importance of each gene to PC 1. In PCA terminology, the weights are called “loadings” and an array of “loadings” for a PC is called an “eigenvector” Gene Influence on PC 1 Eigenvector a high 10 b low 0. 5 c medium 3 d low -0. 2 e high 13 f high -14 … … Stat. Quest: PCA Clearly Explained, by Joshua Starmer, www. seqquest. com “Loadings” or “Weights”
- Slides: 126