SSAHA Search with Speed Nick Altemose Kelvin Gu
- Slides: 1

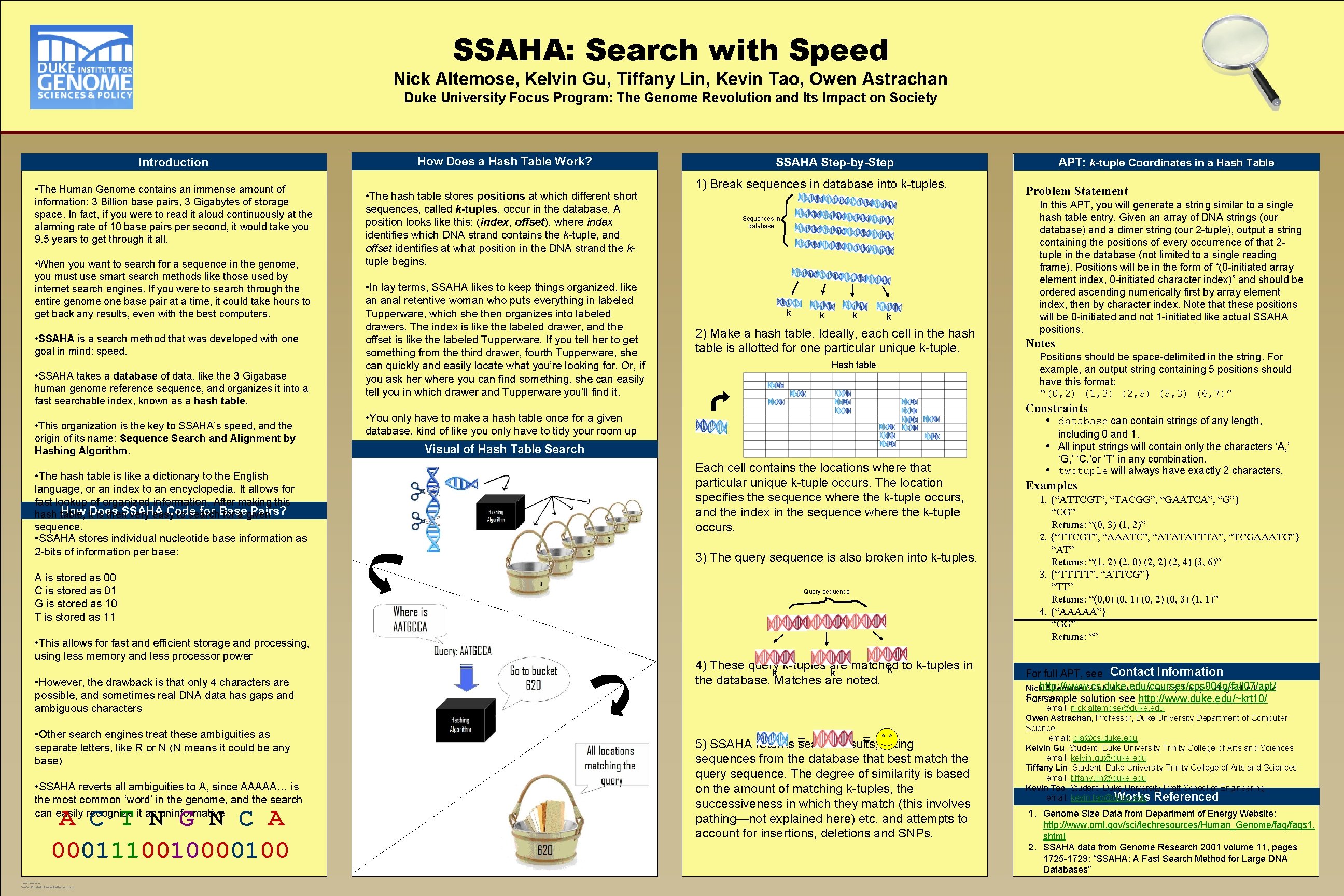
SSAHA: Search with Speed Nick Altemose, Kelvin Gu, Tiffany Lin, Kevin Tao, Owen Astrachan Duke University Focus Program: The Genome Revolution and Its Impact on Society Introduction • The Human Genome contains an immense amount of information: 3 Billion base pairs, 3 Gigabytes of storage space. In fact, if you were to read it aloud continuously at the alarming rate of 10 base pairs per second, it would take you 9. 5 years to get through it all. • When you want to search for a sequence in the genome, you must use smart search methods like those used by internet search engines. If you were to search through the entire genome one base pair at a time, it could take hours to get back any results, even with the best computers. • SSAHA is a search method that was developed with one goal in mind: speed. • SSAHA takes a database of data, like the 3 Gigabase human genome reference sequence, and organizes it into a fast searchable index, known as a hash table. • This organization is the key to SSAHA’s speed, and the origin of its name: Sequence Search and Alignment by Hashing Algorithm. • The hash table is like a dictionary to the English language, or an index to an encyclopedia. It allows for fast lookup of organized information. After making this How it. Does SSAHA Code for Base Pairs? hash table, is then very easy to search for a given sequence. • SSAHA stores individual nucleotide base information as 2 -bits of information per base: A is stored as 00 C is stored as 01 G is stored as 10 T is stored as 11 • This allows for fast and efficient storage and processing, using less memory and less processor power • However, the drawback is that only 4 characters are possible, and sometimes real DNA data has gaps and ambiguous characters • Other search engines treat these ambiguities as separate letters, like R or N (N means it could be any base) • SSAHA reverts all ambiguities to A, since AAAAA… is the most common ‘word’ in the genome, and the search can easily recognize it as uninformative A C T N G N C A 00011100100 TEMPLATE DESIGN © 2007 www. Poster. Presentations. com How Does a Hash Table Work? • The hash table stores positions at which different short sequences, called k-tuples, occur in the database. A position looks like this: (index, offset), where index identifies which DNA strand contains the k-tuple, and offset identifies at what position in the DNA strand the ktuple begins. • In lay terms, SSAHA likes to keep things organized, like an anal retentive woman who puts everything in labeled Tupperware, which she then organizes into labeled drawers. The index is like the labeled drawer, and the offset is like the labeled Tupperware. If you tell her to get something from the third drawer, fourth Tupperware, she can quickly and easily locate what you’re looking for. Or, if you ask her where you can find something, she can easily tell you in which drawer and Tupperware you’ll find it. SSAHA Step-by-Step 1) Break sequences in database into k-tuples. Sequences in database k k 2) Make a hash table. Ideally, each cell in the hash table is allotted for one particular unique k-tuple. Hash table APT: k-tuple Coordinates in a Hash Table Problem Statement In this APT, you will generate a string similar to a single hash table entry. Given an array of DNA strings (our database) and a dimer string (our 2 -tuple), output a string containing the positions of every occurrence of that 2 tuple in the database (not limited to a single reading frame). Positions will be in the form of “(0 -initiated array element index, 0 -initiated character index)” and should be ordered ascending numerically first by array element index, then by character index. Note that these positions will be 0 -initiated and not 1 -initiated like actual SSAHA positions. Notes Positions should be space-delimited in the string. For example, an output string containing 5 positions should have this format: “(0, 2) (1, 3) (2, 5) (5, 3) (6, 7)” Constraints • You only have to make a hash table once for a given database, kind of like you only have to tidy your room up once, then you can find things. Table extremely easily thereafter. Visual of Hash Search • • Each cell contains the locations where that particular unique k-tuple occurs. The location specifies the sequence where the k-tuple occurs, and the index in the sequence where the k-tuple occurs. 3) The query sequence is also broken into k-tuples. Query sequence 4) These query k-tuples are matched to k-tuples in k k k the database. Matches are noted. = listing 5) SSAHA returns = search results, sequences from the database that best match the query sequence. The degree of similarity is based on the amount of matching k-tuples, the successiveness in which they match (this involves pathing—not explained here) etc. and attempts to account for insertions, deletions and SNPs. • database can contain strings of any length, including 0 and 1. All input strings will contain only the characters ‘A, ’ ‘G, ’ ‘C, ’or ‘T’ in any combination. twotuple will always have exactly 2 characters. Examples 1. {“ATTCGT”, “TACGG”, “GAATCA”, “G”} “CG” Returns: “(0, 3) (1, 2)” 2. {“TTCGT”, “AAATC”, “ATATATTTA”, “TCGAAATG”} “AT” Returns: “(1, 2) (2, 0) (2, 2) (2, 4) (3, 6)” 3. {“TTTTT”, “ATTCG”} “TT” Returns: “(0, 0) (0, 1) (0, 2) (0, 3) (1, 1)” 4. {“AAAAA”} “GG” Returns: “” For full APT, see Contact Information Nickhttp: //www. cs. duke. edu/courses/cps 004 g/fall 07/apt/ Altemose, Student, Duke University Trinity College of Arts and Sciences For sample solution see http: //www. duke. edu/~krt 10/ email: nick. altemose@duke. edu Owen Astrachan, Professor, Duke University Department of Computer Science email: ola@cs. duke. edu Kelvin Gu, Student, Duke University Trinity College of Arts and Sciences email: kelvin. gu@duke. edu Tiffany Lin, Student, Duke University Trinity College of Arts and Sciences email: tiffany. lin@duke. edu Kevin Tao, Student, Duke University Pratt School of Engineering email: kevin. tao@duke. edu Works Referenced 1. Genome Size Data from Department of Energy Website: http: //www. ornl. gov/sci/techresources/Human_Genome/faqs 1. shtml 2. SSAHA data from Genome Research 2001 volume 11, pages 1725 -1729: “SSAHA: A Fast Search Method for Large DNA Databases”