Splice sites consensus sequence substitution and Mutation affecting

Splice sites, consensus sequence substitution and Mutation affecting Translation P 3, Classes –VII M. Sc. -IV Semester Dr. Hifzur R Siddique Section of Genetics Department of Zoology ALIGARH MUSLIM UNIVERSITY 1

Splice sites mutation

Introns always have two distinct nucleotides at either end. At the 5' end the DNA nucleotides are GT [GU in the pre-messenger RNA (pre-m. RNA)]; at the 3' end they are AG. These nucleotides are part of the splicing sites. DONOR-SPLICE: splicing site at the beginning of an intron, intron 5' left end. ACCEPTOR-SPLICE: splicing site at the end of an intron, intron 3' right end. A polypyrimidine (Cn. Tn) motif is present upstream of the CAG intron 3' ending. More upstream, the consensus branch site (CTGAC) (not shown) is a necessary component in the effective splicing of the pre-m. RNA. The sn. RP's or "snurps" (small nuclear ribonucleoproteins) search out the sequences above and join together with other sn. RP's to form a Spliceosome. These cut out the introns, forming the "lariat formation" of the excised intron.
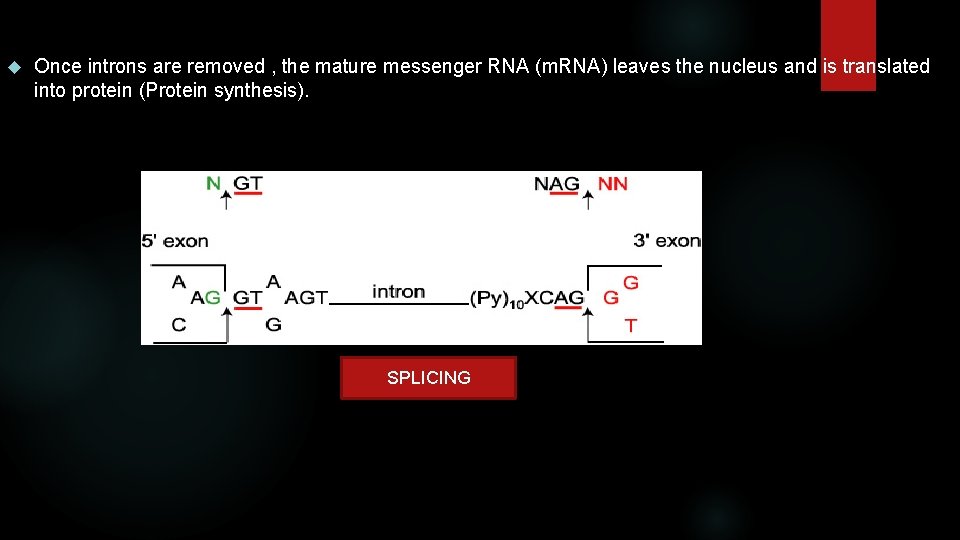
Once introns are removed , the mature messenger RNA (m. RNA) leaves the nucleus and is translated into protein (Protein synthesis). SPLICING

Functional studies of cloned 6 -globin genes have confirmed the necessity of the invariant sequences located at the 5' end of introns for normal RNA processing. Similar studies have also demonstrated that alternative patterns of RNA splicing are revealed by mutations that abolish normal splicing signals or create new splice sites in introns or exons. In Greek and Italian populations, mutations which result in altered globin RNA splicing appear to be common causes of 1 -thalassemia.
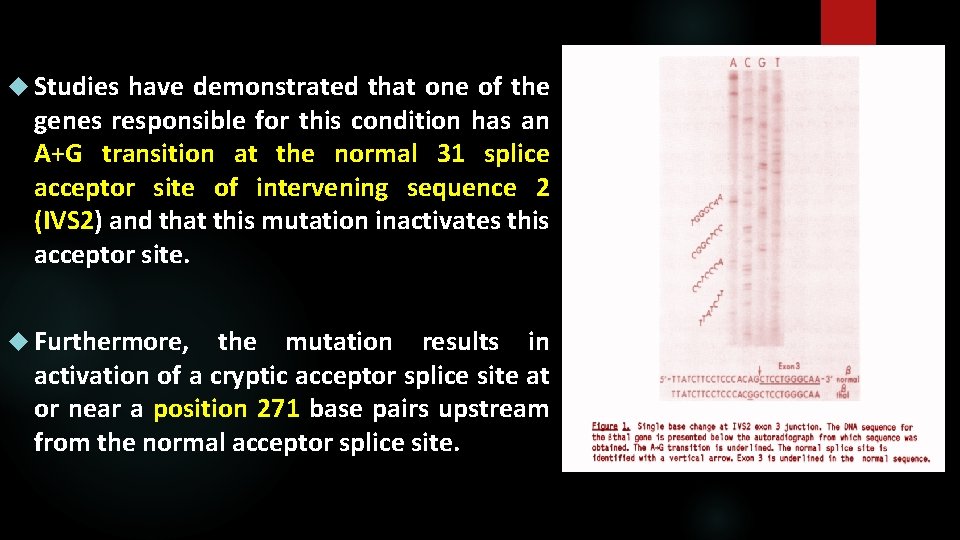
Studies have demonstrated that one of the genes responsible for this condition has an A+G transition at the normal 31 splice acceptor site of intervening sequence 2 (IVS 2) and that this mutation inactivates this acceptor site. Furthermore, the mutation results in activation of a cryptic acceptor splice site at or near a position 271 base pairs upstream from the normal acceptor splice site.

Consensus sequence substitutions

In this schematic representation, eight related TE sequences are shown as thick horizontal lines. Most sequence variations occur at discrete positions, but some are also shared between copies. Changes include single nucleotide substitutions (vertical ticks) and insertions (triangle above the sequence) and deletions (interrupted line) of variable size.

The K. pneumonia of the DNA sequences of majority of nif gene promoters have highly conserved sequence between position 11 and 26 with respect to the transcription initiation. The consensus sequence is TG-N 8 -TTGCA and is quite unlike the consensus sequence found in the promoter of prokaryotes. Within this consensus three residues remain invariant, namely the GG pair at 25, -24 and the G at -13. Modification of residues between -13 and -24 had relatively little effect, causing a reduction of at most 50 percent in promoter activity.

In some cases, mutation in these sequences can actually increase promoter activity. But in general, mutation at -12, -13 -24, -25 has a strong promoter down phenotype. At the -12 position a C to T transition cause marked promoter down phenotype activity in the nif. L promoter but the same mutation is silent in nif. H promoter.

Mutation affecting translation

Mutation that enhance or reduce the efficiency with which ribosomes bind to an m. RNA, could have a large effect on gene expression. Initiation of translation varies greatly from one m. RNA to another. But, there is evidence for a consensus sequence immediately upstream from the initiator AUG. This affects the initiation rate for most of the eukaryotes.

Cont… The sequence is CC(A or G)CCAUG, where the underlined codon is initiator codon. A person homozygous for a deletion of the two nucleotide at the position -2 and -3 prior to the AUG codon is known. This person has alpha-thalassemia, which is caused by very limited translatin of the alphaglobin m. RNA that contain the deletion.

In all organisms there is a preference towards the use of a specific subset of codons, referred to as ‘optimal codons’, which usually constitute 70– 90% of the codons in a gene and match the most abundant t. RNA in the cell. Optimal codons increase the accuracy of t. RNA selection (translation accuracy) and also the total amount of the produced protein (translation efficiency). A possible explanation for these phenomena is that, by using codons that are non‐optimal to the t. RNA pool, Translation Elongation accuracy is reduced due to higher rates of amino acid misincorporation. is slowed down due to increased translational proofreading resulting in lower translation efficiency.
For example, In E. coli, antibiotics such as streptomycin (STR) and spectinomycin (SPEC) were shown to bind to the ribosome and modify the accuracy of translation. Many studies in E. coli have shown that Alterations in the rps. L (the gene encoding the small subunit ribosomal (r) protein S 12) produce streptomycin resistance (STRR) and error‐restrictive ribosomes (i. e. ribosomes with reduced translational error rates). In contrast, mutations in the rps. E (the gene encoding the small subunit r‐protein S 5) produce spectinomycin resistance (SPECR) and error‐prone ribosomes (i. e. ribosomes with increased translational error rates)

Two common rps. L mutations in E. coli that confer STRR are the replacement of the amino acid lysine 42 with threonine (referred to as K 42 T) and the replacement of the amino acid lysine 87 with arginine (referred to as K 87 R). While the K 42 T mutation also results in error‐restrictive ribosomes, the K 87 R mutation results in semi error‐restrictive ribosomes, with a translational error rate that is lower than that of wild‐type strain yet higher than error‐restrictive strains such as the K 42 T mutant. On the other hand, it was demonstrated that a single amino acid substitution in the E. coli rps. E gene of a highly conserved glycine to an aspartate at position 28 (referred to G 28 D), which produces SPECR, also resulted in error‐prone ribosomes.

Position of mutations in S 12 and S 5 proteins of E. coli K‐ 12. The position numbering originates from the start codon of the open reading frame.

h T k n a Y u o
- Slides: 18