Specimen Data Refinery A Landscape Analysis on Machine










- Slides: 23

Specimen Data Refinery A Landscape Analysis on Machine Learning, Computer Vision and Automated Approaches to Create Specimen Metadata 23 -10 -2019 Biodiversity_Next Laurence Livermore 1 & Rob Cubey 2 Natural History Museum, London, UK 2. Royal Botanic Garden Edinburgh, UK https: //doi. org/10. 3897/biss. 3. 37647 1.

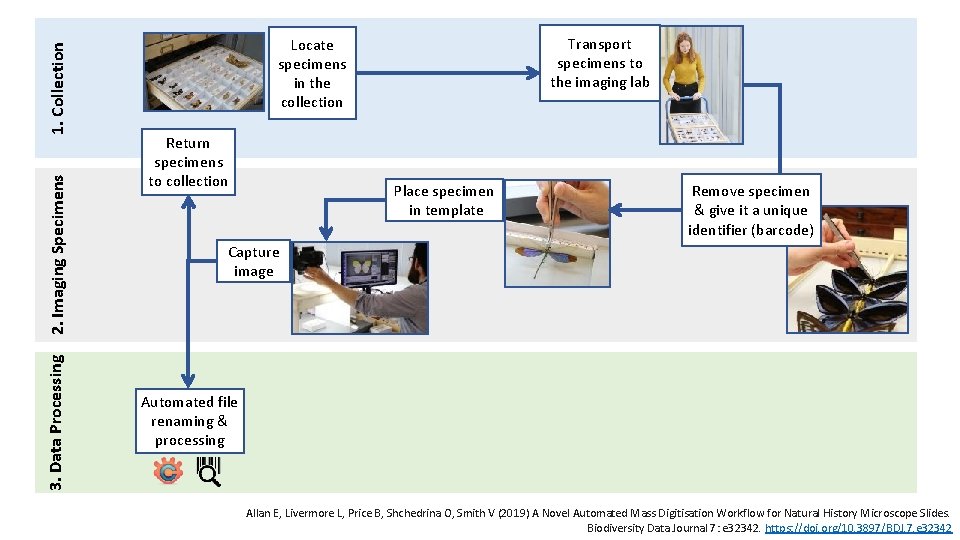
1. Collection 2. Imaging Specimens 3. Data Processing Transport specimens to the imaging lab Locate specimens in the collection Return specimens to collection Place specimen in template Remove specimen & give it a unique identifier (barcode) Capture image Automated file renaming & processing Allan E, Livermore L, Price B, Shchedrina O, Smith V (2019) A Novel Automated Mass Digitisation Workflow for Natural History Microscope Slides. Biodiversity Data Journal 7: e 32342. https: //doi. org/10. 3897/BDJ. 7. e 32342

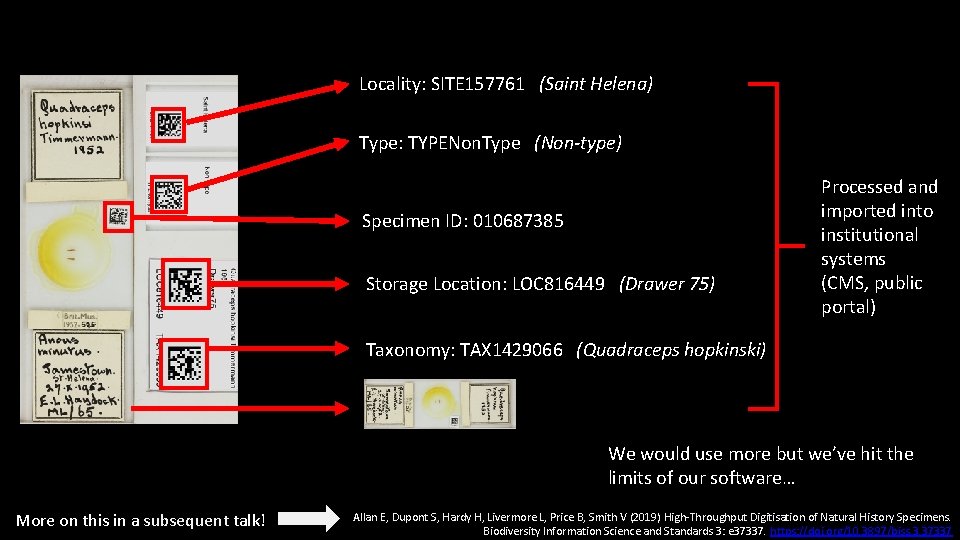
Locality: SITE 157761 (Saint Helena) Type: TYPENon. Type (Non-type) Specimen ID: 010687385 Storage Location: LOC 816449 (Drawer 75) Processed and imported into institutional systems (CMS, public portal) Taxonomy: TAX 1429066 (Quadraceps hopkinski) We would use more but we’ve hit the limits of our software… More on this in a subsequent talk! Allan E, Dupont S, Hardy H, Livermore L, Price B, Smith V (2019) High-Throughput Digitisation of Natural History Specimens. Biodiversity Information Science and Standards 3: e 37337. https: //doi. org/10. 3897/biss. 3. 37337

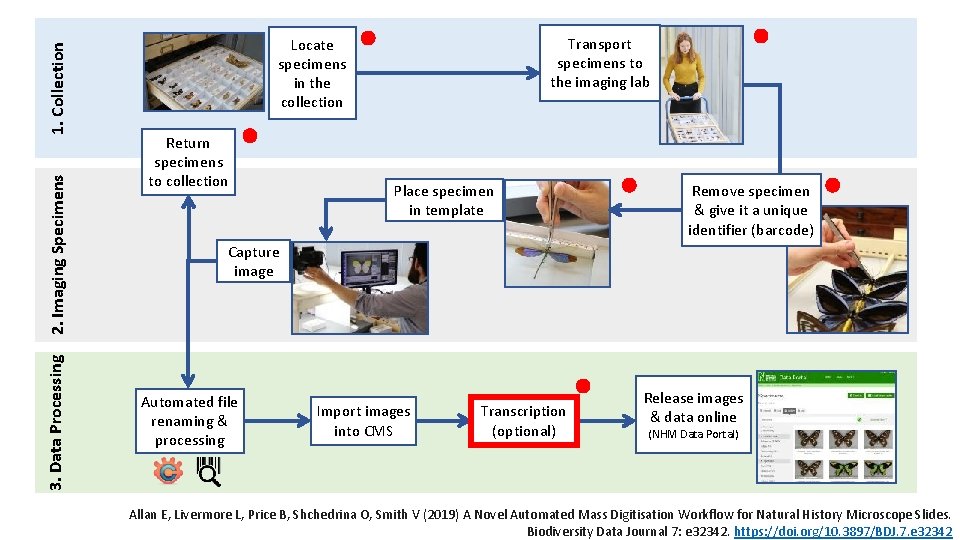
1. Collection 2. Imaging Specimens 3. Data Processing Transport specimens to the imaging lab Locate specimens in the collection Return specimens to collection Place specimen in template Remove specimen & give it a unique identifier (barcode) Capture image Automated file renaming & processing Import images into CMS Transcription (optional) Release images & data online (NHM Data Portal) Allan E, Livermore L, Price B, Shchedrina O, Smith V (2019) A Novel Automated Mass Digitisation Workflow for Natural History Microscope Slides. Biodiversity Data Journal 7: e 32342. https: //doi. org/10. 3897/BDJ. 7. e 32342



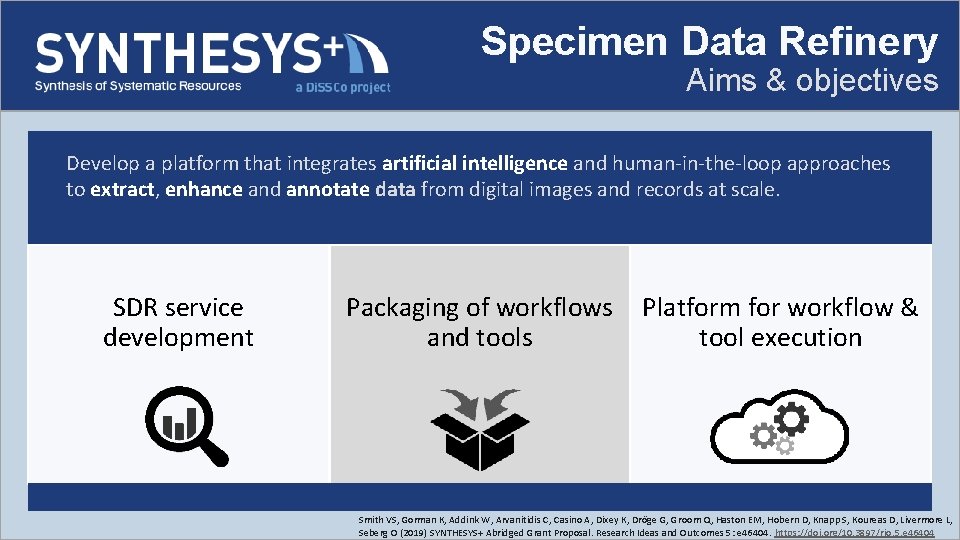
Specimen Data Refinery Aims & objectives Develop a platform that integrates artificial intelligence and human-in-the-loop approaches to extract, enhance and annotate data from digital images and records at scale. SDR service development Packaging of workflows and tools Platform for workflow & tool execution Smith VS, Gorman K, Addink W, Arvanitidis C, Casino A, Dixey K, Dröge G, Groom Q, Haston EM, Hobern D, Knapp S, Koureas D, Livermore L, Seberg O (2019) SYNTHESYS+ Abridged Grant Proposal. Research Ideas and Outcomes 5: e 46404. https: //doi. org/10. 3897/rio. 5. e 46404

Easier • Condition checking (e. g. gum chloral/phenol balsam discoloration, verdigris, pyrite oxidation) • Natural language descriptions of specimens (e. g. for public, curators, researchers) Microscope slide with gum chloral discoloration This is a caterpillar of the common green birdwing. We acquired it in 1939. It is part of the Lionel Walter Rothschild bequeathment in our entomological collections. It dimensions are 9. 5 cm long. • Taxonomic trait extraction (e. g. phenology, morphology, biological relationships) Harder Slide image courtesy of Zoe Jay Adams (NHM)

Task 8. 1 Landscape evaluation • Evaluate platform based approaches inc. tool/service registries • Identify sources of data • Identify & create training/reference datasets • Assess potential to use EUDAT, EGI, EOSC, AWS etc • Reuse & integrate reports from previous Di. SSCorelated work Task 8. 2 Development of tools, services & workflows • Services include: • Optical character recognition data capture and analysis • Data mining and linkage • Image analysis • Georeferencing • Human interaction and crowdsourcing Mature services will be containerized and incorporated into SDR workflow & tool registry Summary of Tasks Task 8. 3 Development of SDR cloud platform • Platform will run workflows in SDR registry • Controlled user access (ELIXIR authentication & authorisation) • Create detailed standardized provenance metadata of derivative datasets • Derivative datasets and outputs packaged as Research Objects (ROs) • Ledger log of all ROs & specimen UIDs Task 8. 4 Data delivery & exploitation • User acceptance testing (against Di. SSCo user stories) • Training and workshops to promote use • Delivery of data to collections management systems • Exploitation of data by third parties

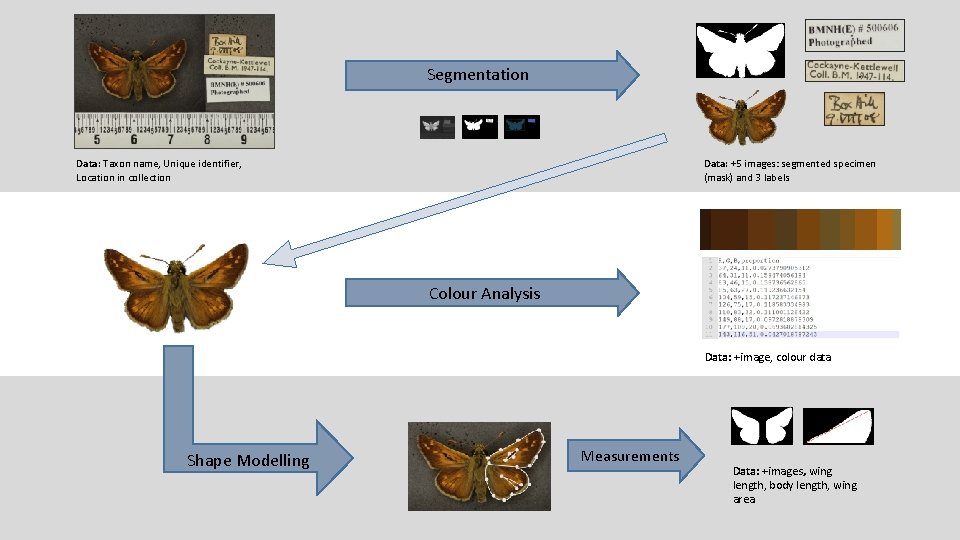
Segmentation Data: Taxon name, Unique identifier, Location in collection Data: +5 images: segmented specimen (mask) and 3 labels Colour Analysis Data: +image, colour data Shape Modelling Measurements Data: +images, wing length, body length, wing area

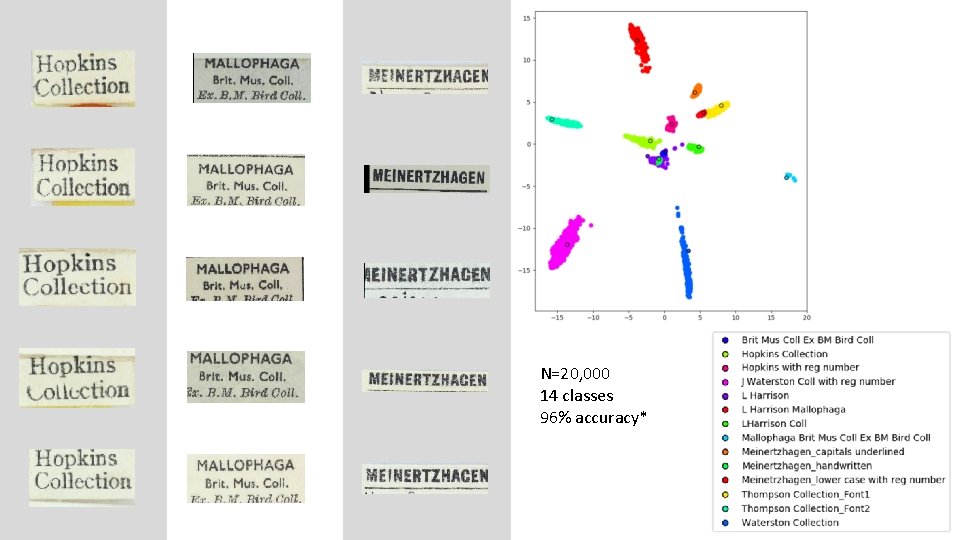
N=20, 000 14 classes 96% accuracy*

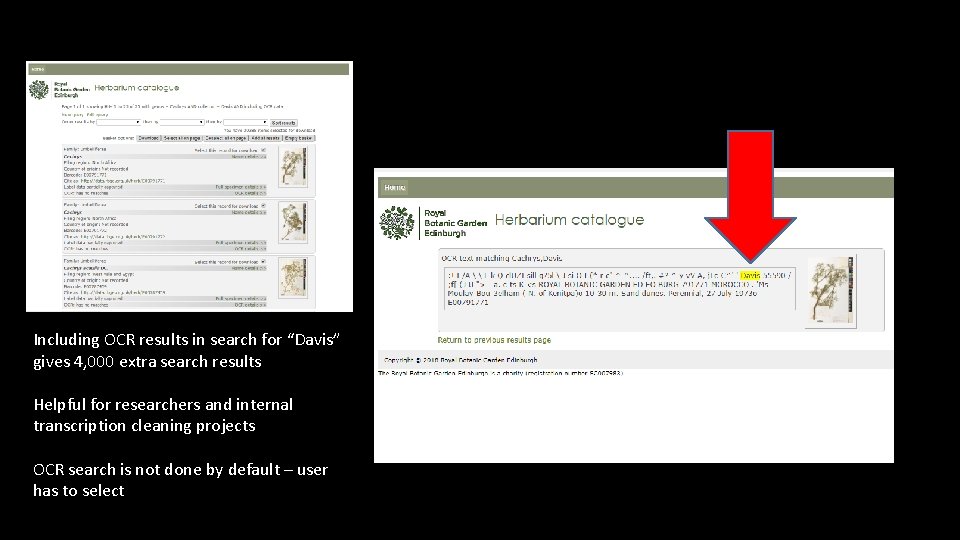
Fide'pbvis "n Plortx op Turkey ; 1: 30 S (rtb^) ROYAL BOTANIC GARDEN EDINBURGH E 00374073 Davis & Hedge (D. 31208 A). I atui. �Ji^Crac. A f M m *�m Turkey. Pr ov�Tunceli: xmzur dag above Q^aoik* 24. 00 m. Screes. Perennial � 16 <, J� 957 � E 00374073 ABBYY Finereader > Reads whole herbarium sheet images > generates unparsed text

Including OCR results in search for “Davis” gives 4, 000 extra search results Helpful for researchers and internal transcription cleaning projects OCR search is not done by default – user has to select


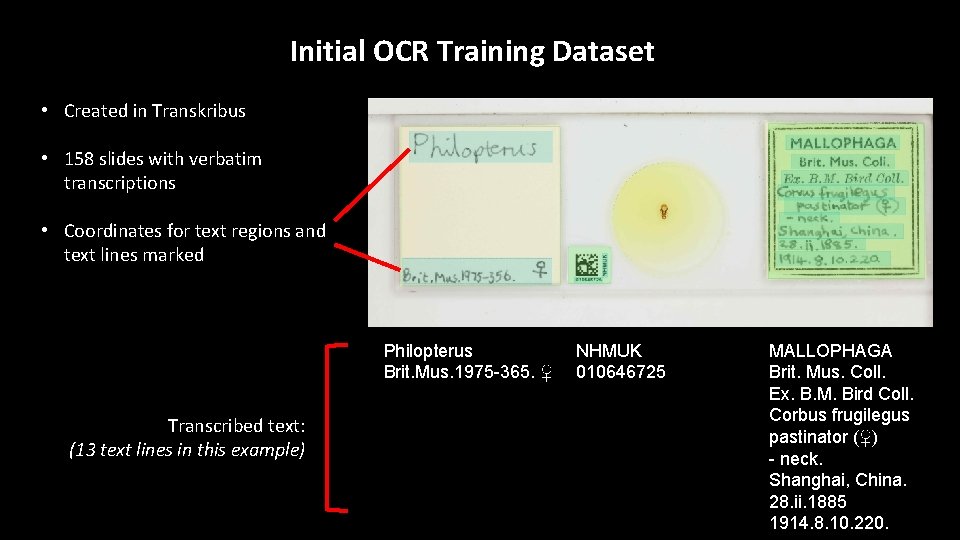
Initial OCR Training Dataset • Created in Transkribus • 158 slides with verbatim transcriptions • Coordinates for text regions and text lines marked Philopterus Brit. Mus. 1975 -365. ♀ Transcribed text: (13 text lines in this example) NHMUK 010646725 MALLOPHAGA Brit. Mus. Coll. Ex. B. M. Bird Coll. Corbus frugilegus pastinator (♀) - neck. Shanghai, China. 28. ii. 1885 1914. 8. 10. 220.

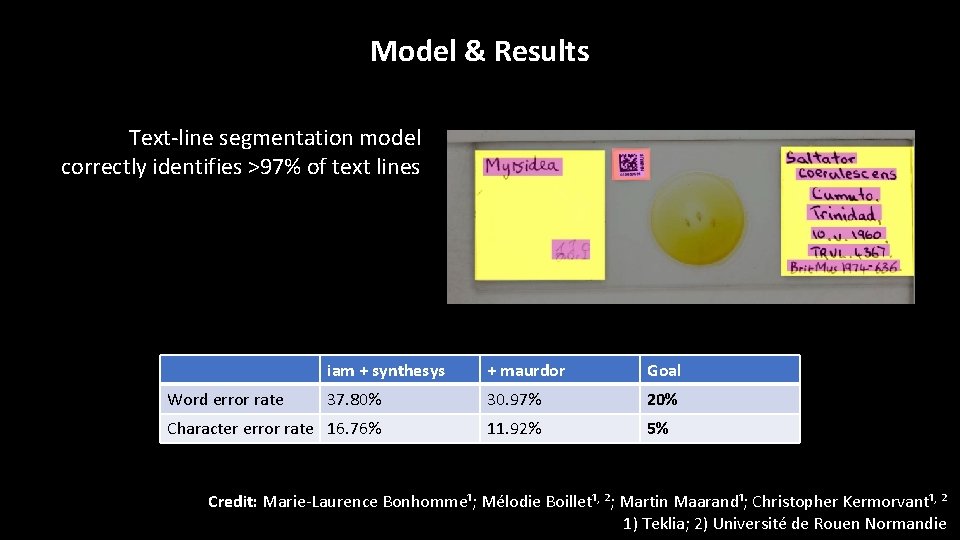
Model & Results Text-line segmentation model correctly identifies >97% of text lines iam + synthesys + maurdor Goal 37. 80% 30. 97% 20% Character error rate 16. 76% 11. 92% 5% Word error rate Credit: Marie-Laurence Bonhomme¹; Mélodie Boillet¹, ²; Martin Maarand¹; Christopher Kermorvant¹, ² 1) Teklia; 2) Université de Rouen Normandie

Expected: Rajputana. March Error: Rapulana. March Expected: Lucknow ( U. P. ) Error: Lucknown ( U. P. )

Labels Spacers Type Label Barcode Label Specimen/Coverslip


More on segmentation in next talk! Abraham Nieva de la Hidalga et al Use of Semantic Segmentation for Increasing the Throughput of Digitisation Workflows for Natural History Collections. https: //doi. org/10. 3897/biss. 3. 37161

Next Steps Now • Collect more user stories • Create larger OCR datasets • Expand pilot to other collection types (herb sheets, pinned insects) • Create object detection training datasets for instance segmentation • Discuss approaches to national/local gazetteer services • Agree on common data standard(s) and tool/service versioning • Trial creating/storing provenance data / ways of communicating epistemology 1+ year • Trial and deploy formally described workflows on a cloud platform


Publications & Datasets • Methods for automated text digitisation (Owen et al, 2019) • Dillen M, Groom Q, Chagnoux S, Güntsch A, Hardisty A, Haston E, Livermore L, Runnel V, Schulman L, Willemse L, Wu Z, Phillips S (2019) A benchmark dataset of herbarium specimen images with label data. Biodiversity Data Journal 7: e 31817. https: //doi. org/10. 3897/BDJ. 7. e 31817 • Smith VS, Gorman K, Addink W, Arvanitidis C, Casino A, Dixey K, Dröge G, Groom Q, Haston EM, Hobern D, Knapp S, Koureas D, Livermore L, Seberg O (2019) SYNTHESYS+ Abridged Grant Proposal. Research Ideas and Outcomes 5: e 46404. https: //doi. org/10. 3897/rio. 5. e 46404 • Allan E, Dupont S, Hardy H, Livermore L, Price B, Smith V (2019) High-Throughput Digitisation of Natural History Specimens. Biodiversity Information Science and Standards 3: e 37337. https: //doi. org/10. 3897/biss. 3. 37337 • Phillips Sarah, Dillen Mathias, Groom Quentin, Green Laura, Weech Marie- Hélène, & Wijkamp Noortje. (2019). Report on New Methods for Data Quality Assurance, Verification and Enrichment. Zenodo. https: //doi. org/10. 5281/zenodo. 3364509 • Quentin Groom, Mathias Dillen, Helen Hardy, Sarah Phillips, Luc Willemse, & Zhengzhe Wu. (2019). Updating data standards in transcription. Zenodo. https: //doi. org/10. 5281/zenodo. 3386211 • Bug. Snapper: Camera Design for High-throughput Digitisation (Mark Hereld, Argonne National Laboratory) • Open. Refine Platform (Stan. DAP-Herb) (Fabian Reimeier, BGBM) SYNTHESYS 3 • Report: Automating data capture from natural history specimens • Report: Digitisation on Demand • Software: Inselect – desktop application for NH image processing, barcode reading, metadata