SPECIES IDENTIFICATION THROUGH DNA STRING ANALYSIS Mark Vorster










![Research Overview - Bioinformatics - String Matching - Discussion - Questions Example Grouping Seq[336] Research Overview - Bioinformatics - String Matching - Discussion - Questions Example Grouping Seq[336]](https://slidetodoc.com/presentation_image/1d4d0b97797c9ce1d9febb199c9f1a4d/image-15.jpg)

- Slides: 17

SPECIES IDENTIFICATION THROUGH DNA STRING ANALYSIS Mark Vorster Supervisor: Prof Philip Machanick

Research Overview - Bioinformatics - String Matching - Discussion - Questions Research Overview Goal Aid bioinformaticians in research by providing a tool which can identify similar DNA sequences in order to infer homogeneity, in a timely manner. Reason for problems Large data sets Days of processing No existing specific tools 2

Research Overview - Bioinformatics - String Matching - Discussion - Questions Bioinformatics "Research, development, or application of computational tools and approaches for expanding the use of biological, medical, behavioural or health data, including those to acquire, store, organize, archive, analyse, or visualise such data. “ Biomedical Information Science and Technology Initiatives Definition Committee - Dr Huerta "The branch of science concerned with information and information flow in biological systems, esp. the use of computational methods in genetics and genomics. “ Oxford English Dictionary 3

Research Overview - Bioinformatics - String Matching - Discussion - Questions History of Bioinformatics and Genetics 1953 - Watson, Crick , Wilkins and Franklin. One helical turn = 3. 4 nm Discrete abstraction Adenine – Thymine Guanine – Cytosine Sugar-phosphate backbone base Hydrogen bonds http: //www. accessexcellence. org/RC/VL/GG/images/structure. gif 4

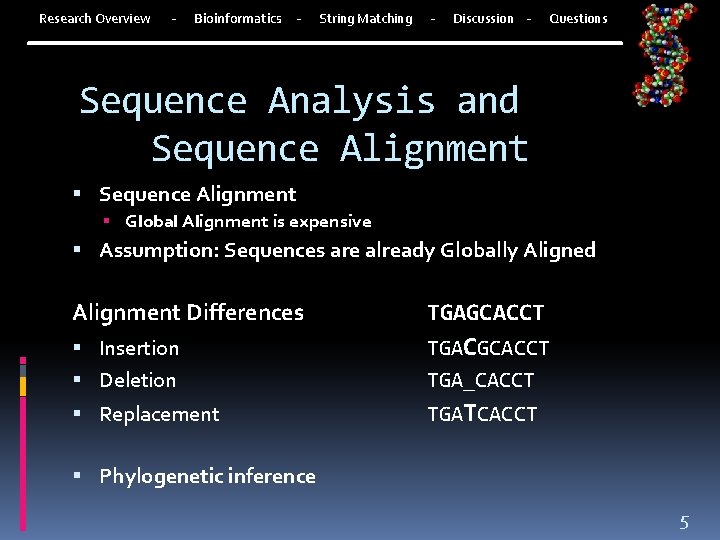
Research Overview - Bioinformatics - String Matching - Discussion - Questions Sequence Analysis and Sequence Alignment Global Alignment is expensive Assumption: Sequences are already Globally Aligned Alignment Differences TGAGCACCT Insertion Deletion TGACGCACCT TGA_CACCT Replacement TGATCACCT Phylogenetic inference 5

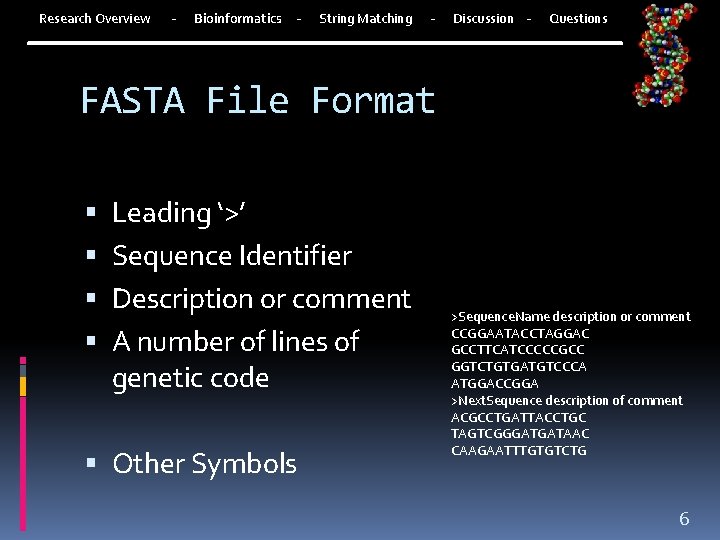
Research Overview - Bioinformatics - String Matching - Discussion - Questions FASTA File Format Leading ‘>’ Sequence Identifier Description or comment A number of lines of genetic code Other Symbols >Sequence. Name description or comment CCGGAATACCTAGGAC GCCTTCATCCCCCGCC GGTCTGTGATGTCCCA ATGGACCGGA >Next. Sequence description of comment ACGCCTGATTACCTGC TAGTCGGGATGATAAC CAAGAATTTGTGTCTG 6

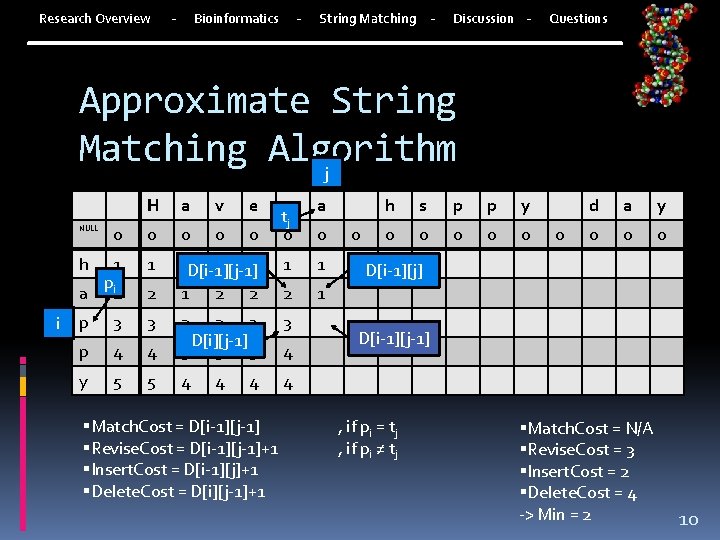
Research Overview - Bioinformatics - String Matching - Discussion - Questions Approximate String Matching Algorithm Nesting loops inefficient Dynamic Programing Take into account all previous information Improved to O(n 2) | where n is number of bases in shorter sequence Goal: Find the closet match between two strings Or the minimum number of differences 7

Research Overview - Bioinformatics - String Matching - Discussion - Questions Approximate String Matching Algorithm Minimum of: Match. Cost = D[i-1][j-1] , if pi = tj Revise. Cost = D[i-1][j-1]+1 , if pi ≠ tj Insert. Cost = D[i-1][j]+1 D[i-1][j-1] Delete. Cost = D[i][j-1]+1 D[i][j-1] D[i-1][j] D[i][j] D[0][j] = 0 and D[i][0] = i 8

Research Overview - - Bioinformatics - String Matching Discussion - Questions Approximate String Matching Algorithm NULL 0 h 1 a 2 p 3 p 4 y 5 H a v e 0 0 a 0 0 0 h s p p y 0 0 0 d a y 0 0 0 9

Research Overview - - Bioinformatics - String Matching Discussion - Questions Approximate String Matching Algorithm j H a v e 0 0 0 1 p a 2 i 1 1 1 2 1 D[i-1][j-1] 1 1 1 2 2 2 1 p 3 3 2 3 p 4 4 2 3 D[i][j-1] 3 3 3 y 5 5 4 4 NULL h i 4 4 Match. Cost = D[i-1][j-1] Revise. Cost = D[i-1][j-1]+1 Insert. Cost = D[i-1][j]+1 Delete. Cost = D[i][j-1]+1 tj 0 4 a 0 0 h s p p y 0 0 0 d a y 0 0 0 D[i-1][j] D[i-1][j-1] , if pi = tj , if pi ≠ tj Match. Cost = N/A Revise. Cost = 3 Insert. Cost = 2 Delete. Cost = 4 -> Min = 2 10

Research Overview - - Bioinformatics - String Matching Discussion - Questions Approximate String Matching Algorithm H a v e a h s p p y NULL 0 0 0 0 h 1 1 1 1 0 1 1 a 2 2 1 2 1 1 2 2 2 p 3 3 2 2 2 2 1 2 3 p 4 4 3 3 3 3 2 1 2 y 5 5 4 4 4 4 3 2 1 0 d a y 0 0 0 11

Research Overview - Bioinformatics - String Matching - Discussion - Questions Approximate String Matching Algorithm Changes D[i][0] = i , if pi = t 0 D[i][0] = i + 1 , if pi ≠ t 0 D[0][j] = j , if p 0 = tj D[0][j] = j + 1 , if p 0 ≠ tj Additional stop case for mismatch 12

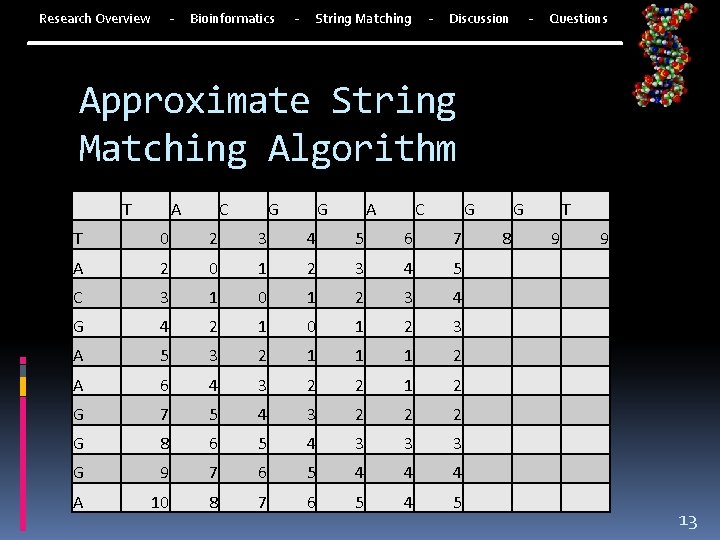
- Research Overview Bioinformatics - - String Matching - Discussion Questions Approximate String Matching Algorithm T A C G G A C G T 0 2 3 4 5 6 7 A 2 0 1 2 3 4 5 C 3 1 0 1 2 3 4 G 4 2 1 0 1 2 3 A 5 3 2 1 1 1 2 A 6 4 3 2 2 1 2 G 7 5 4 3 2 2 2 G 8 6 5 4 3 3 3 G 9 7 6 5 4 4 4 A 10 8 7 6 5 4 5 G 8 T 9 9 13

Research Overview - Bioinformatics - String Matching - Discussion - Questions Discussion Grouping Algorithm Scale of the problem 400 – 800 bases per sequence Ten thousands of sequences Assumptions: Sequences Globally Aligned Sequences Begin at the Same Place 14
![Research Overview Bioinformatics String Matching Discussion Questions Example Grouping Seq336 Research Overview - Bioinformatics - String Matching - Discussion - Questions Example Grouping Seq[336]](https://slidetodoc.com/presentation_image/1d4d0b97797c9ce1d9febb199c9f1a4d/image-15.jpg)
Research Overview - Bioinformatics - String Matching - Discussion - Questions Example Grouping Seq[336] HK 2 QS 7 R 01 AXRJ 6 Seq[218] Seq[38] Seq[235] Seq[89] … Seq[382] HK 2 QS 7 R 01 BR 4 Q 9 Seq[173] Seq[180] HK 2 QS 7 R 01 ABFDP Seq[339] Seq[289] Seq[491] Seq[319] … Seq[269] HK 2 QS 7 R 01 AZHD 7 Seq[402] Seq[112] Seq[203] Seq[137] … Seq[210] HK 2 QS 7 R 01 BMNQ 4 Seq[364] Seq[270] HK 2 QS 7 R 01 AZFOG Seq[388] Seq[441] Seq[442] HK 2 QS 7 R 01 ADASO Seq[426] Seq[233] Seq[374] Seq[416] … … … 15

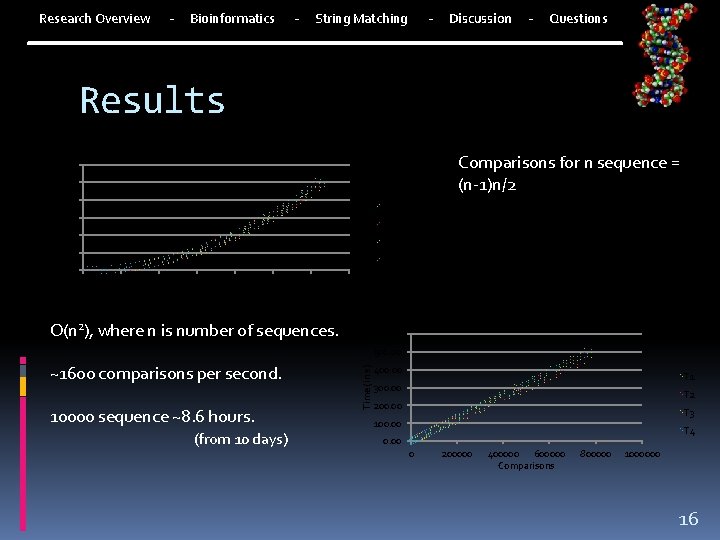
- Research Overview Bioinformatics - - String Matching Discussion - Questions Results Time to complete for X Sequences Comparisons for n sequence = (n-1)n/2 600. 00 400. 00 T 1 300. 00 T 2 200. 00 T 3 100. 00 T 4 0. 00 0 200 400 600 800 Sequences 1000 1200 1400 Time to complete for X Comparisons O(n 2), where n is number of sequences. 600. 00 500. 00 ~1600 comparisons per second. 10000 sequence ~8. 6 hours. (from 10 days) Time (in s) 500. 00 400. 00 T 1 300. 00 T 2 200. 00 T 3 100. 00 T 4 0. 00 0 200000 400000 600000 Comparisons 800000 1000000 16

Research Overview - Bioinformatics - String Matching - Discussion ? - Questions