Speciation l The actual origin of species l



























- Slides: 40


Speciation l The actual “origin of species” l Reduction in gene flow, genetic and phenotypic change in populations l The study of speciation requires that species be real

Speciation l Speciation is the antidote to sex l Keeps together adaptive groups of traits l New species most often uniparental l Phylogeny is genealogy of species l Branching tree

Hybridization l Mules l Fertile interspecific hybrids are common in perennial plants

Hybrid speciation l New species form from interspecific hybrids l Two parents l Phylogenetic pattern is reticulate l Enough examples to make it interesting l Not enough to disrupt the generally divergent pattern of phylogeny

Alloploidy l Chromosome doubling (unreduced gametes or somatic doubling) l Alloploid effectively has one diploid set from each parent species l Instant Speciation™

Homoploid hybrid speciation l No chromosome doubling l Two theoretical modes, both documented § Recombinational speciation § Speciation with external barriers

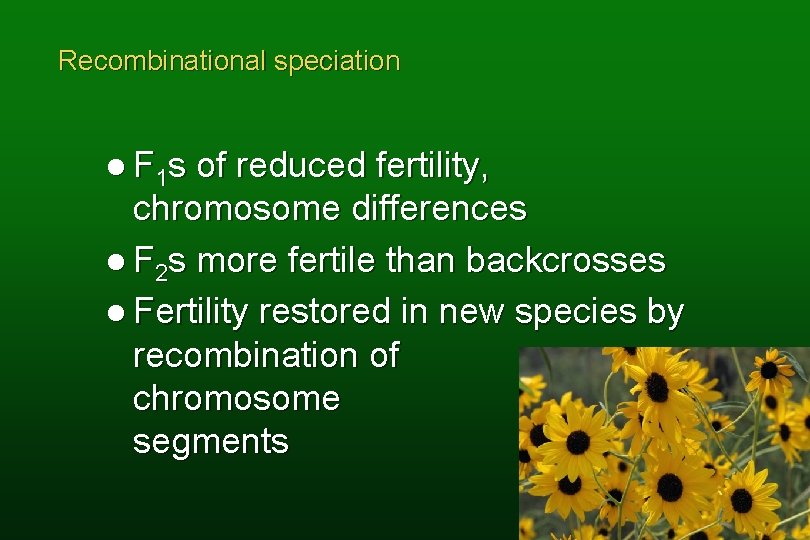
Recombinational speciation l F 1 s of reduced fertility, chromosome differences l F 2 s more fertile than backcrosses l Fertility restored in new species by recombination of chromosome segments

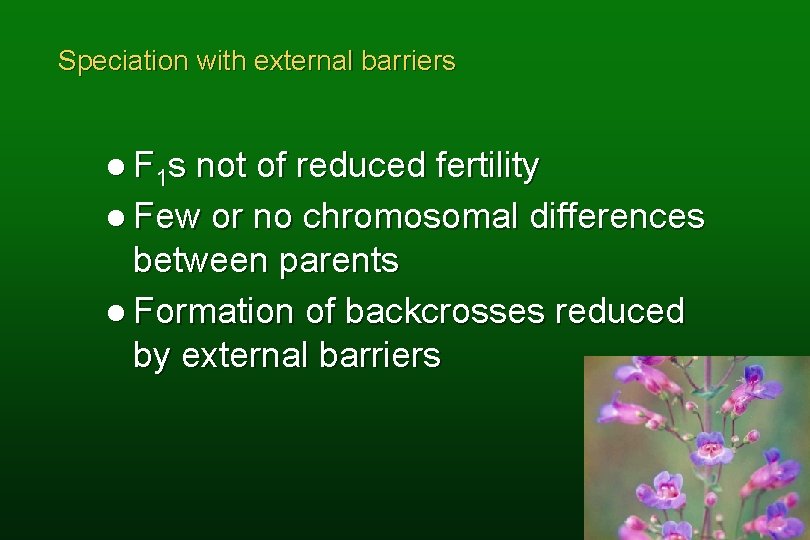
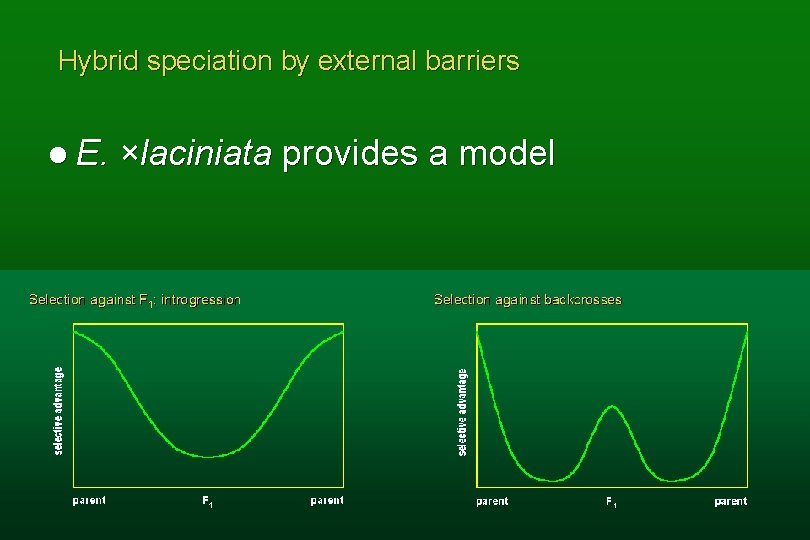
Speciation with external barriers l F 1 s not of reduced fertility l Few or no chromosomal differences between parents l Formation of backcrosses reduced by external barriers

A long time ago, in a desert close at hand…

So what’s an Encelia? l Asteraceae (sunflower family) l Mostly shrubs l Dry habitats, mostly deserts l Brittlebush (E. farinosa)

A hybrid under every bush l “The bushes are hybrids” l All species are interfertile l No apparent reduction of fertility in F 1, F 2, backcross l Is it a syngameon?

Spontaneous natural hybrids 1. 2. 3. 4. 5. 6. 7. 8. 9. E. farinosa × E. frutescens E. farinosa × E. californica E. farinosa × E. palmeri E. farinosa × E. halimifolia E. californica × E. asperifolia E. ventorum × E. palmeri E. ventorum × E. asperifolia E. virginensis × E. frutescens E. actoni × E. frutescens

Encelia ×laciniata Named as a species l Hybrids between E. ventorum and E. palmeri l Selection against recombinants l

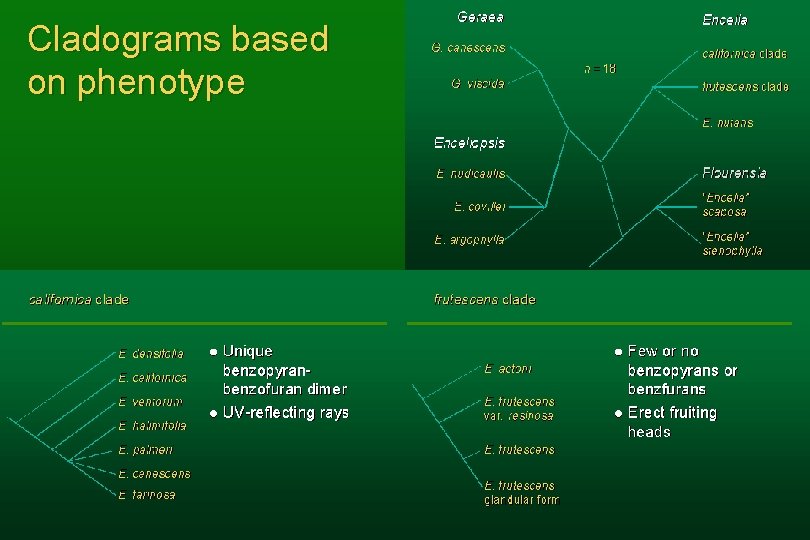
Phylogeny: always a good place to start l The days of cladistics before DNA l Two well-defined clades (californica clade and frutescens clade) l Relationships within clades less clear

Not just morphology—phenotype Standard morphology of heads, capitulescences, leaves l Micromorphology, especially trichomes l Secondary chemistry l Ultraviolet floral patterns l Anatomy of stems and leaves (petioles turned out to be useful) l More that I’ve probably forgotten l

Cladograms based on phenotype

DNA sequence analysis l ITS (internal transcribed spacer of ribosomal DNA) l DNA doesn’t work so well for closely related species l Hybridization is more likely to be confusing in DNA sequence analysis than in morphological analysis


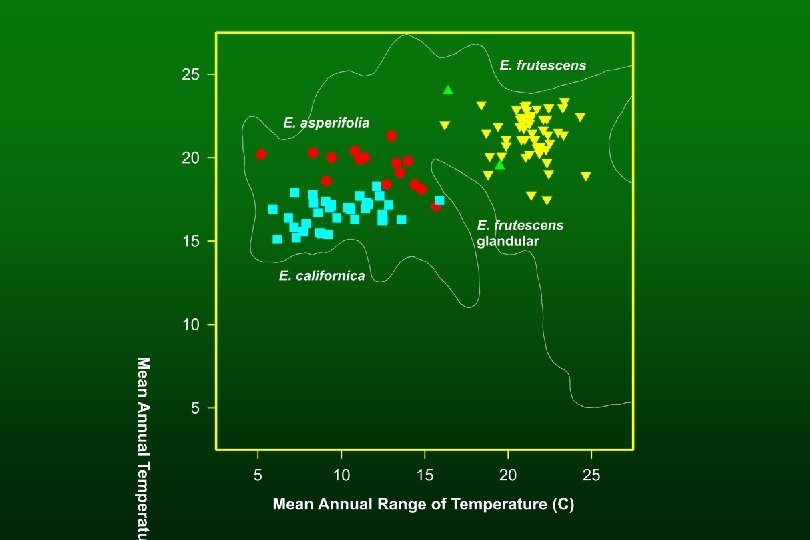
Identifying species of hybrid origin l Species of hybrid origin not always intermediate between parents l Species of intermediate morphology not always of hybrid origin

Preponderance of evidence l Intermediate morphology l Agreement with F 1 s l Apomorphies shared with parents

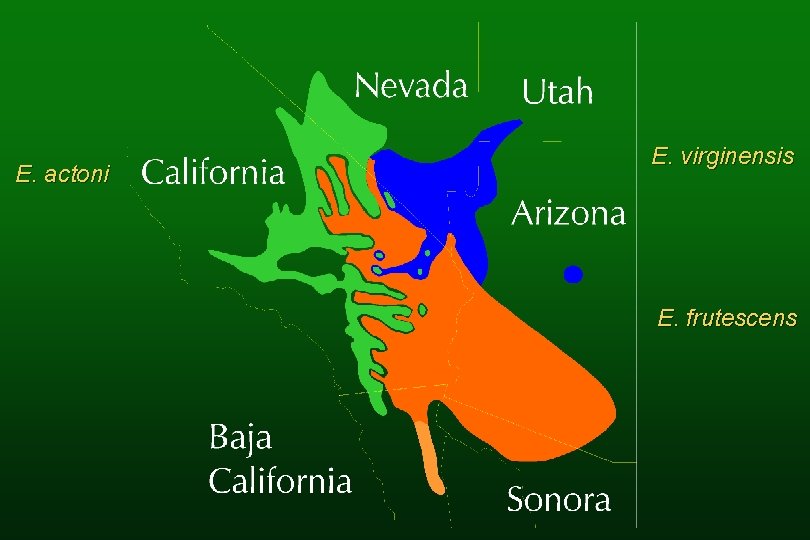
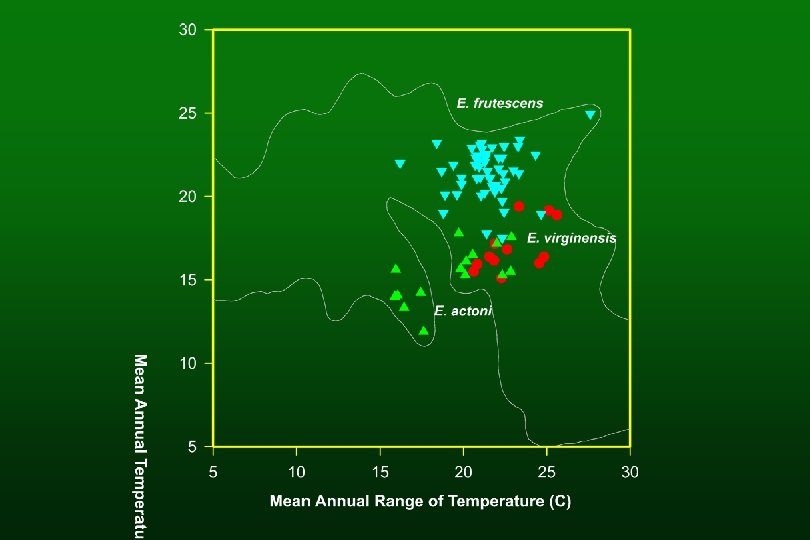
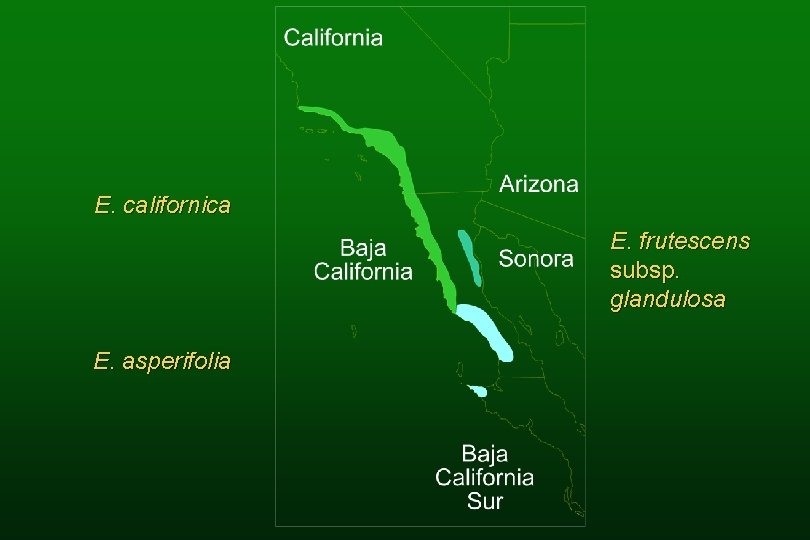
Species of hybrid origin E. virginensis (parents: E. actoni and E. frutescens subsp. frutescens) E. asperifolia (parents: E. californica and E. frutescens subsp. glandulosa)

Encelia virginensis

E. actoni E. virginensis E. frutescens

Shared phenotypic apomorphies – E. virginensis l With E. frutescens § broad multicellular-based hairs l With E. actoni § none (E. actoni has no clear autapomorphies, but E. virginensis resembles it morphologically)

Agreement with F 1

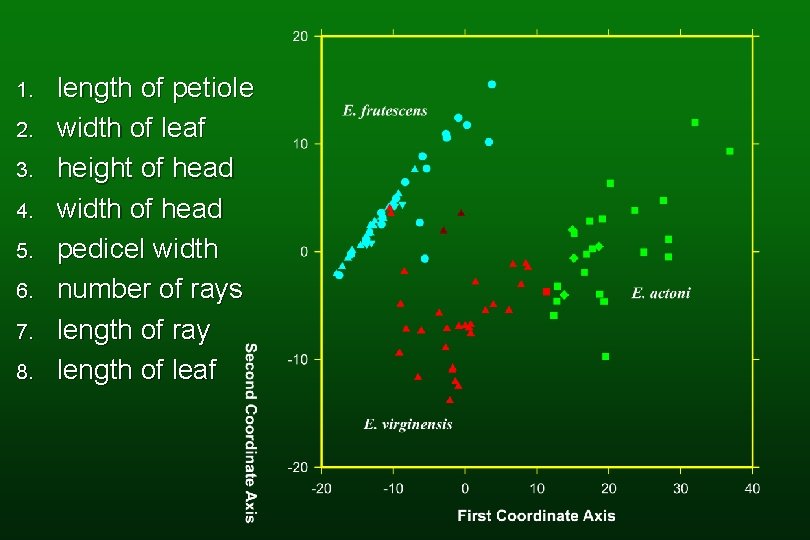
1. 2. 3. 4. 5. 6. 7. 8. length of petiole width of leaf height of head width of head pedicel width number of rays length of ray length of leaf



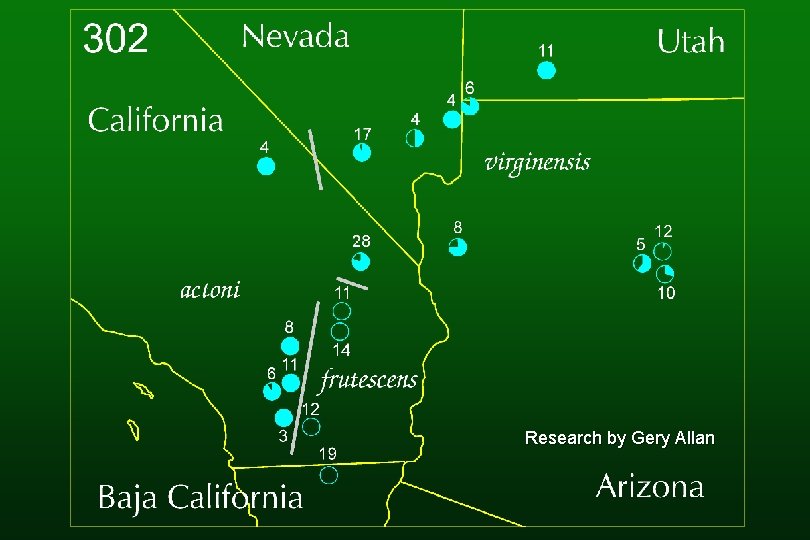
Research by Gery Allan

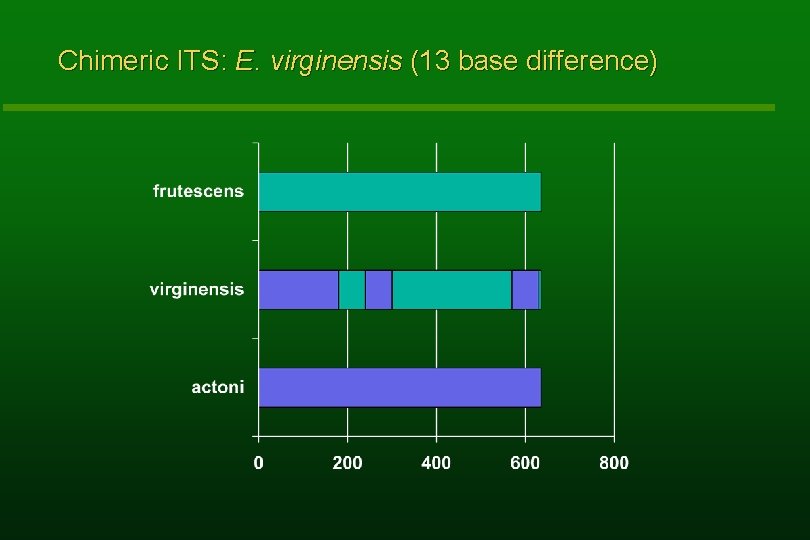
Chimeric ITS: E. virginensis (13 base difference)

Encelia asperifolia

E. californica E. frutescens subsp. glandulosa E. asperifolia

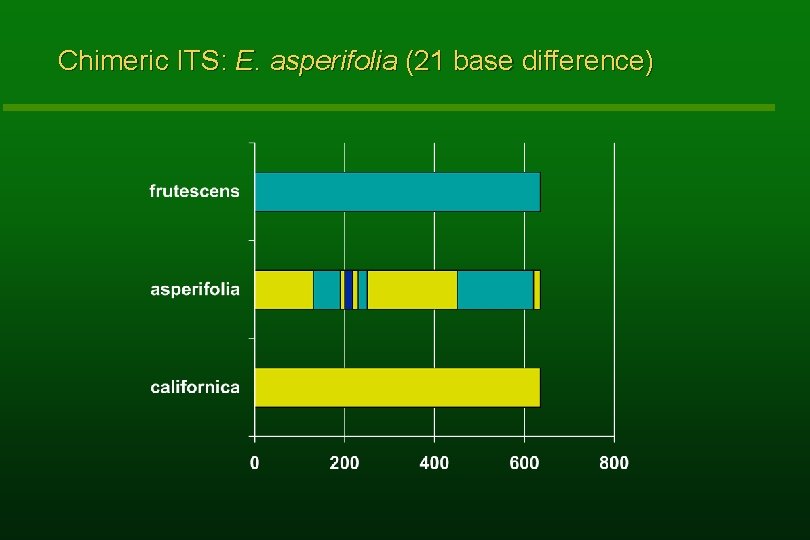
Shared phenotypic apomorphies – E. asperifolia With E. frutescens § broad multicellular-based hairs § no benzopyrans or benzofurans § yellow stigmas l With E. californica § UV-reflective ray corollas § brown disk corollas § moniliform hairs l


RAPD data: E. asperifolia l Shared § § § with E. californica UBC 218 (0. 8 kbase, 1. 6 kbase) UBC 382 (1. 4 kbase) UBC 409 (0. 5 kbase) UBC 478 (1. 4 kbase) Operon B 8 (0. 75 kbase) l Shared with E. frutescens § UBC 149 (0. 7 kbase) § UBC 375 (1. 0 kbase)

Chimeric ITS: E. asperifolia (21 base difference)

Hybrid speciation by external barriers l E. ×laciniata provides a model

Conclusion I’m done l What are the traits that adapt the new species to their new habitats? l Are there transgressive traits? l Plenty of other plant genera l

Acknowledgments Allan, Gery J. Axelrod, Daniel Braden, Gerald Bryant, Stephen Budzikiewicz, Herbert Carpenter, Kevin J. Charest, Nancy A. Clark, Emily Ehleringer, James R. Harrington, Daniel F. Isman, Murray B. Kinney, Michael Koukol, Scott R. Kyhos, Donald W. Lahmeyer, Sean C. Laufenberg, Gabriela Lee, Gregory J. Maepo, Linda Miller, David Nishida, Joy H. Panero, José Parra, Mima Patterson, Mark Politt, Ursula Proksch, Peter Rieseberg, Loren Rodriguez, Eloy Saccoman, Stephanie Sanders, Donald L. Schilling, Edward Thompson, William C. Weiler, Jeff Weisman, Kathy Wisdom, Charles Wollenweber, Eckhard Wray, Victor
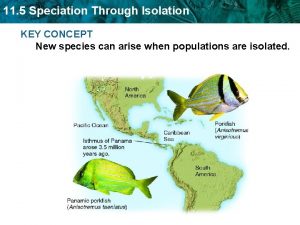
 Sympatric speciation vs allopatric speciation
Sympatric speciation vs allopatric speciation Speciation, or the formation of new species, is
Speciation, or the formation of new species, is Theoretical yeild
Theoretical yeild Sanctifying grace vs actual grace
Sanctifying grace vs actual grace Keystone species definition biology
Keystone species definition biology The origin of species bl
The origin of species bl Origin of species ch 22
Origin of species ch 22 The origin of species chap 24
The origin of species chap 24 The origin of species chapter 18 manga
The origin of species chapter 18 manga The origin of species chapter 24
The origin of species chapter 24 The origin of species ch 24 manhwa
The origin of species ch 24 manhwa Origin of species chapter 7
Origin of species chapter 7 The origin of species manga 24
The origin of species manga 24 Example of biological species
Example of biological species Habitat isolation
Habitat isolation Origin of species by charles darwin
Origin of species by charles darwin Parapatric speciation
Parapatric speciation Ecological speciation
Ecological speciation Sympatric speciation example
Sympatric speciation example Speciation
Speciation Sympatric speciation
Sympatric speciation Speciation
Speciation La spéciation
La spéciation 16-3 the process of speciation
16-3 the process of speciation Reproductive isolation
Reproductive isolation Speciation
Speciation Speciation
Speciation Speciation process
Speciation process Section 16-3 the process of speciation
Section 16-3 the process of speciation Speciation ecureuil antilope
Speciation ecureuil antilope Factors affecting gene frequency slideshare
Factors affecting gene frequency slideshare Parallel evolution
Parallel evolution Section 16-3 the process of speciation answer key
Section 16-3 the process of speciation answer key Allopatric
Allopatric Speciation process
Speciation process Example of biological species
Example of biological species Speciation
Speciation Speciation process
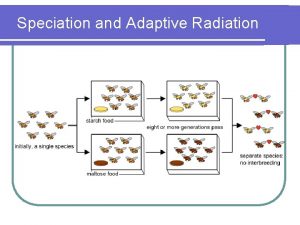
Speciation process Is adaptive radiation divergent evolution
Is adaptive radiation divergent evolution Embryo development
Embryo development Canidae cladogram
Canidae cladogram