Special Topics in Genomics Motif Analysis Sequence motif


- Slides: 13

Special Topics in Genomics Motif Analysis

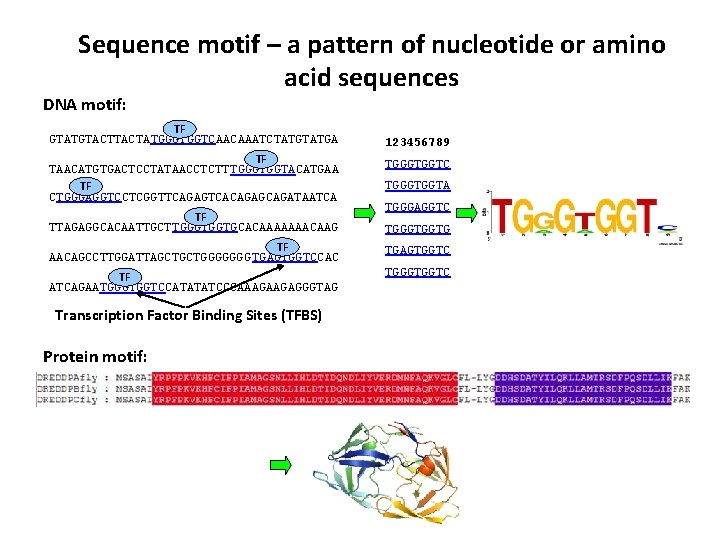
Sequence motif – a pattern of nucleotide or amino acid sequences DNA motif: TF GTATGTACTATGGGTGGTCAACAAATCTATGA TF TAACATGTGACTCCTATAACCTCTT TGGGTGGTACATGAA TF CTGGGAGGTCCTCGGTTCAGAGTCACAGAGCAGATAATCA TF TTAGAGGCACAATTGCTTGGGTGGTGCACAAAAAAACAAG 123456789 TGGGTGGTC TGGGTGGTA TGGGAGGTC TGGGTGGTG TF AACAGCCTTGGATTAGCTGCTGGGGGGG TGAGTGGTCCAC TGAGTGGTC TF ATCAGAATGGGTGGTCCATATATCCCAAAGAAGAGGGTAG TGGGTGGTC Transcription Factor Binding Sites (TFBS) Protein motif:

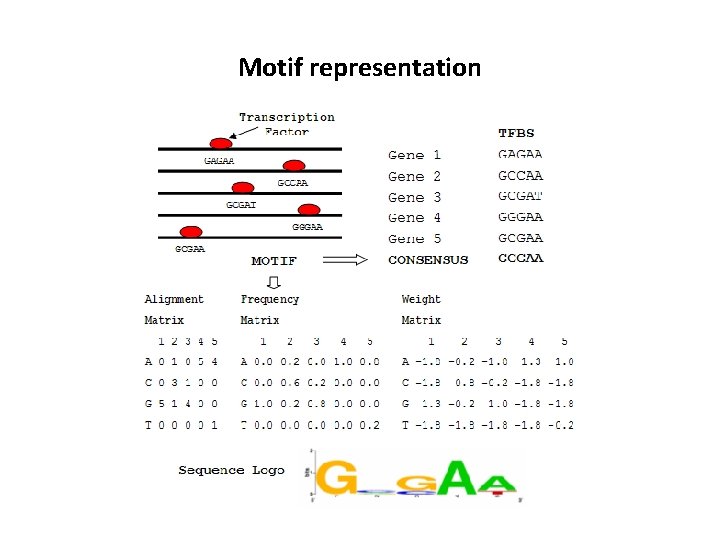
Motif representation

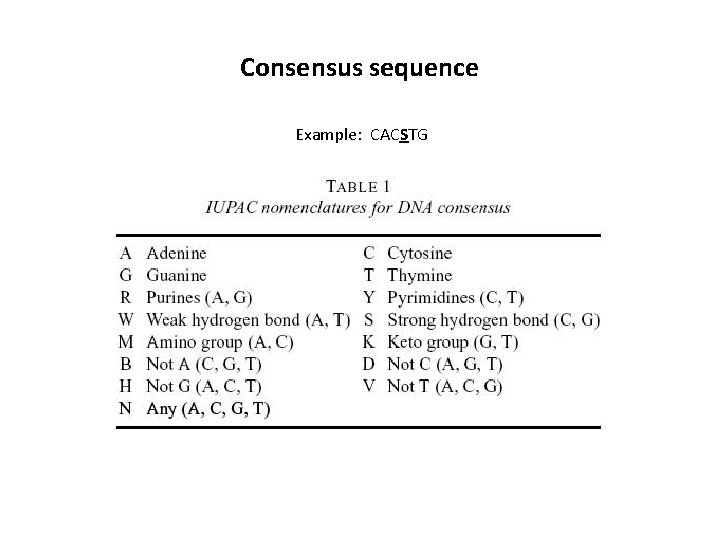
Consensus sequence Example: CACSTG

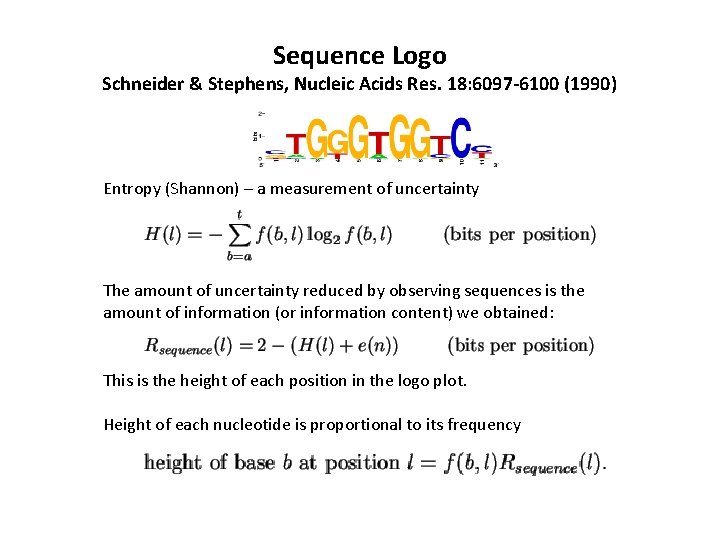
Sequence Logo Schneider & Stephens, Nucleic Acids Res. 18: 6097 -6100 (1990) Entropy (Shannon) – a measurement of uncertainty The amount of uncertainty reduced by observing sequences is the amount of information (or information content) we obtained: This is the height of each position in the logo plot. Height of each nucleotide is proportional to its frequency

Two questions in motif analysis • Known motif mapping Finding occurrences of a motif in nucleotide or amino acid sequences • De novo motif discovery Finding motifs that are previously unknown

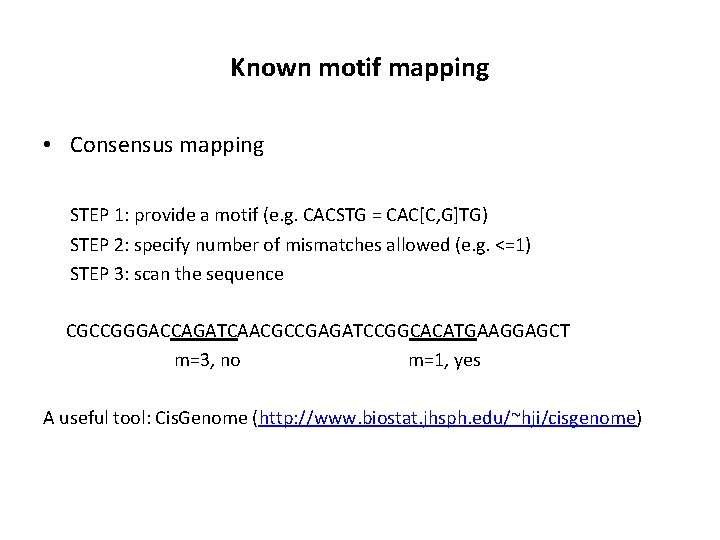
Known motif mapping • Consensus mapping STEP 1: provide a motif (e. g. CACSTG = CAC[C, G]TG) STEP 2: specify number of mismatches allowed (e. g. <=1) STEP 3: scan the sequence CGCCGGGACCAGATCAACGCCGAGATCCGGCACATGAAGGAGCT m=3, no m=1, yes A useful tool: Cis. Genome (http: //www. biostat. jhsph. edu/~hji/cisgenome)

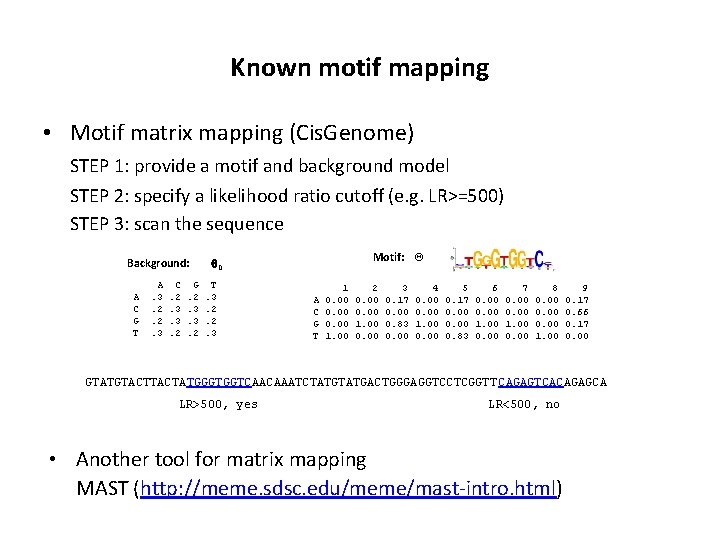
Known motif mapping • Motif matrix mapping (Cis. Genome) STEP 1: provide a motif and background model STEP 2: specify a likelihood ratio cutoff (e. g. LR>=500) STEP 3: scan the sequence Background: A C G T A. 3. 2. 2. 3 C. 2. 3. 3. 2 G. 2. 3. 3. 2 Motif: Q 0 T. 3. 2. 2. 3 A C G T 1 0. 00 1. 00 2 0. 00 1. 00 0. 00 3 0. 17 0. 00 0. 83 0. 00 4 0. 00 1. 00 0. 00 5 0. 17 0. 00 0. 83 6 0. 00 1. 00 0. 00 7 0. 00 1. 00 0. 00 8 0. 00 1. 00 9 0. 17 0. 66 0. 17 0. 00 GTATGTACTATGGGTGGTCAACAAATCTATGACTGGGAGGTCCTCGGTT CAGAGTCACAGAGCA LR>500, yes LR<500, no • Another tool for matrix mapping MAST (http: //meme. sdsc. edu/meme/mast-intro. html)

De novo motif discovery • Two major class of methods: 1. Word enumeration 2. Matrix updating

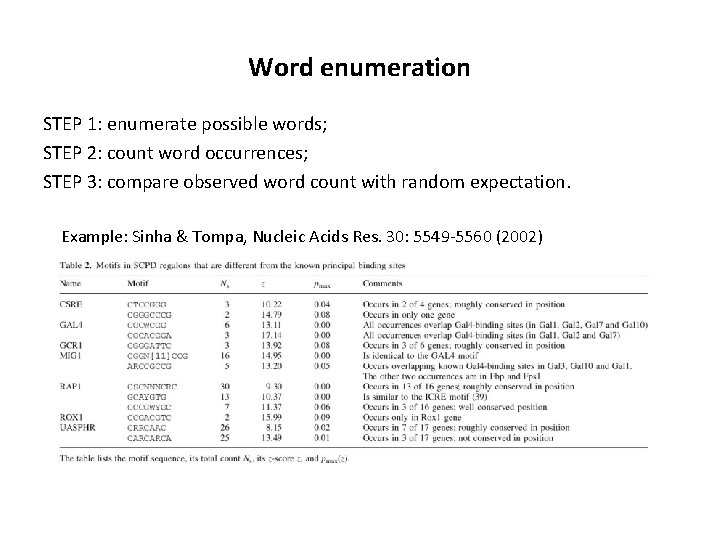
Word enumeration STEP 1: enumerate possible words; STEP 2: count word occurrences; STEP 3: compare observed word count with random expectation. Example: Sinha & Tompa, Nucleic Acids Res. 30: 5549 -5560 (2002)

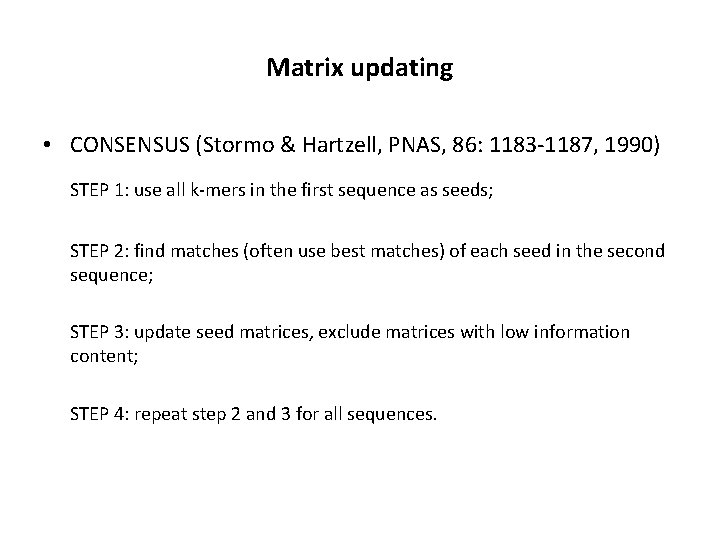
Matrix updating • CONSENSUS (Stormo & Hartzell, PNAS, 86: 1183 -1187, 1990) STEP 1: use all k-mers in the first sequence as seeds; STEP 2: find matches (often use best matches) of each seed in the second sequence; STEP 3: update seed matrices, exclude matrices with low information content; STEP 4: repeat step 2 and 3 for all sequences.

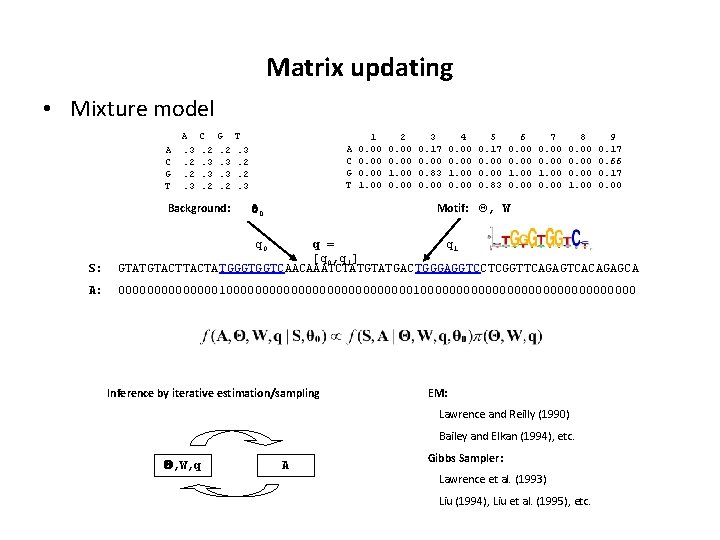
Matrix updating • Mixture model A C G T A. 3. 2. 2. 3 C. 2. 3. 3. 2 G. 2. 3. 3. 2 Background: T. 3. 2. 2. 3 A C G T 1 0. 00 1. 00 2 0. 00 1. 00 0. 00 3 0. 17 0. 00 0. 83 0. 00 4 0. 00 1. 00 0. 00 5 0. 17 0. 00 0. 83 6 0. 00 1. 00 0. 00 7 0. 00 1. 00 0. 00 8 0. 00 1. 00 9 0. 17 0. 66 0. 17 0. 00 Motif: Q, W 0 q 0 S: q = q 1 [q 0, q 1] GTATGTACTATGGGTGGTCAACAAATCTATGACTGGGAGGTCCTCGGTTCAGAGTCACAGAGCA A: 000000000000001000000000000000000000 Inference by iterative estimation/sampling EM: Lawrence and Reilly (1990) Bailey and Elkan (1994), etc. , W, q A Gibbs Sampler: Lawrence et al. (1993) Liu (1994), Liu et al. (1995), etc.

Other issues • Dependencies within motif • Functions of novel motifs