Sorghum Germplasm Mini Core Collection and Genomics for


















- Slides: 50

Sorghum Germplasm Mini Core Collection and Genomics for Enhanced use Hari D Upadhyaya Presented at Genomics for Genebanks (G 4 G) workshop 5 -9 December 2016, CIMMYT, Mexico

Sorghum • 5 th most important cereal crop grown for food, feed, fodder and bioenergy purposes • A staple for over 500 million resource-poor people in marginal environments. • Sorghum production during 2014 was 67. 87 mt from 44. 20 m ha (FAOSTAT, 18 Nov 2016) • USA, Mexico, Nigeria, Sudan and India are the top five sorghum producing countries

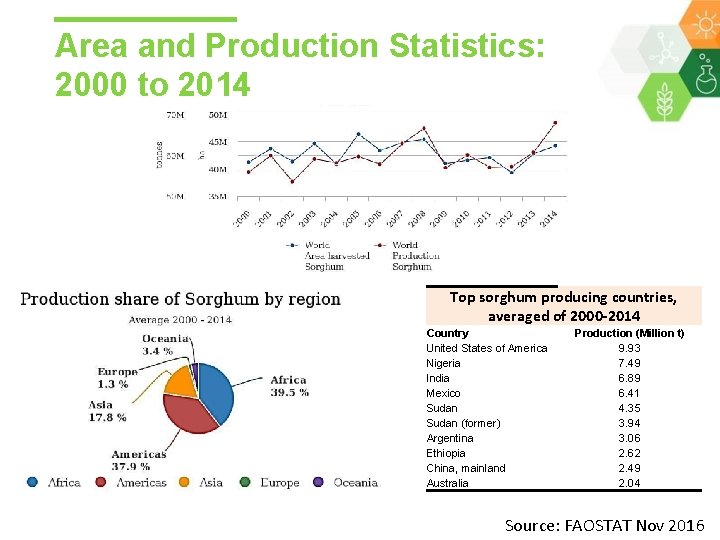
Area and Production Statistics: 2000 to 2014 Top sorghum producing countries, averaged of 2000 -2014 Country United States of America Nigeria India Mexico Sudan (former) Argentina Ethiopia China, mainland Australia Production (Million t) 9. 93 7. 49 6. 89 6. 41 4. 35 3. 94 3. 06 2. 62 2. 49 2. 04 Source: FAOSTAT Nov 2016

Origin and domestication • Domesticated in Ethiopia and surrounding countries, commencing around 4000 -3000 BC, from its wild ancestor Sorghum bicolor subsp. Arundinaceum • Numerous landraces created through of disruptive selection • Spread via the movement of people and trade routes into other regions of Africa, India (Approx. 1500 -1000 BC), the Middle East (approx. 900 – 700 BC) , Far East (approx. 400 AD), and to America (late 1800 s to early 1900 s) (Dillon et al. 2007) • Initial domestication: wild types with small, shattering seed to improved types with larges, non-shattering seed • Three independent mutations in the Sh 1 gene, which encodes a YABBY transcription factor, gave rise to the non-shattering seed phenotype in domesticated sorghum (Olsen 2012; Nature Genetics, 44(6): 616 -617)

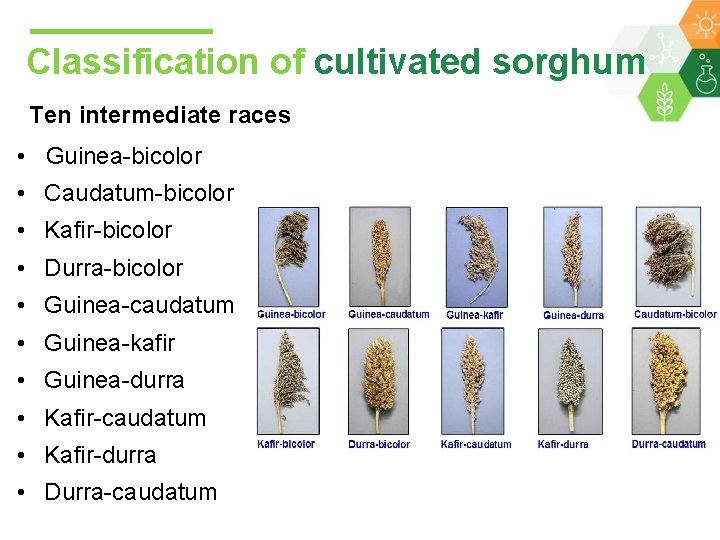
Classification of cultivated sorghum • Sorghum germplasm is classified into five basic races and ten intermediate races (Harlan and de Wet 1972. ) Five basic races : Bicolor Guinea Caudatum Kafir Durra

Classification of cultivated sorghum Ten intermediate races • Guinea-bicolor • Caudatum-bicolor • Kafir-bicolor • Durra-bicolor • Guinea-caudatum • Guinea-kafir • Guinea-durra • Kafir-caudatum • Kafir-durra • Durra-caudatum

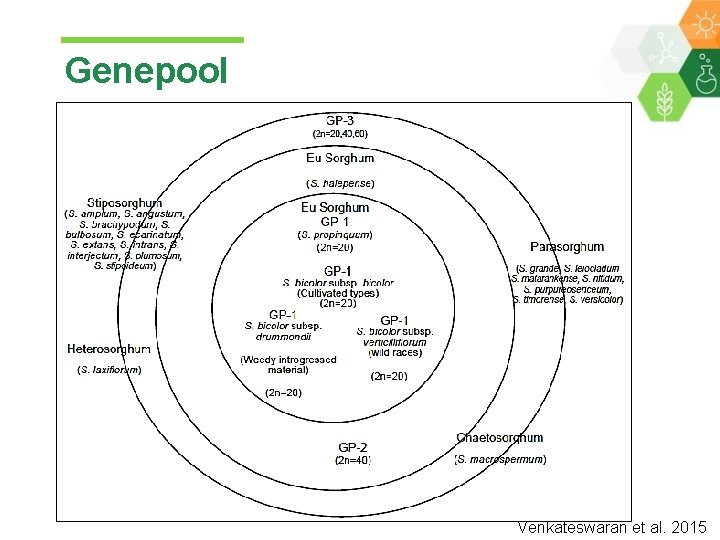
Genepool Venkateswaran et al. 2015

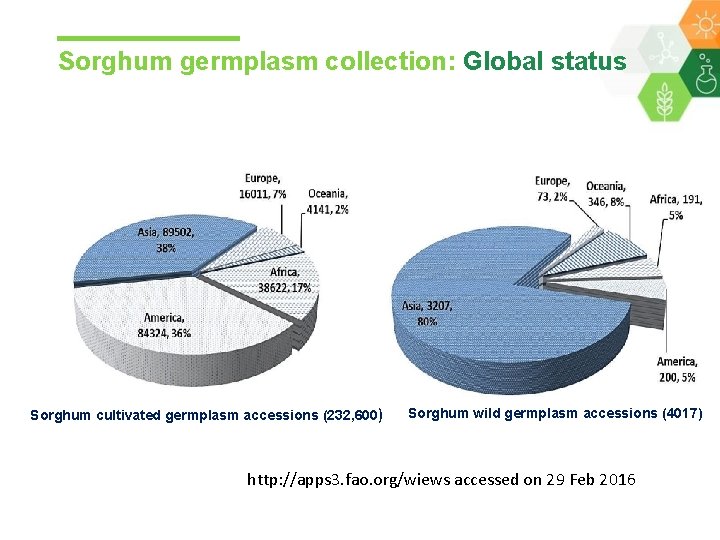
Sorghum germplasm collection: Global status Sorghum cultivated germplasm accessions (232, 600) Sorghum wild germplasm accessions (4017) http: //apps 3. fao. org/wiews accessed on 29 Feb 2016

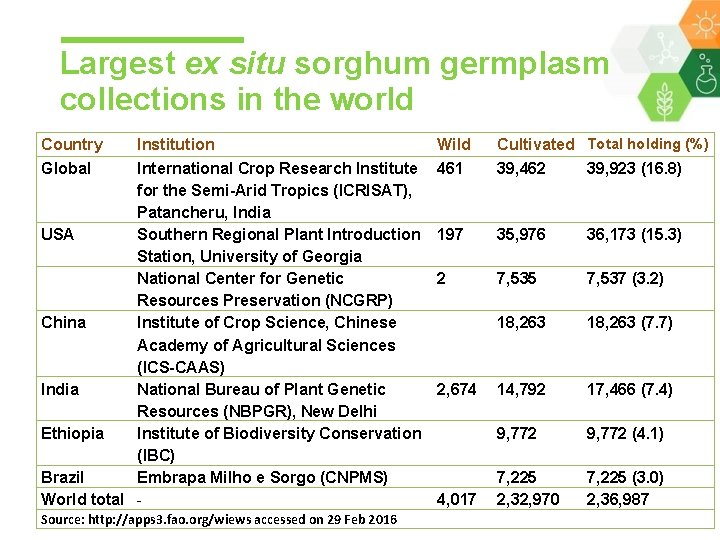
Largest ex situ sorghum germplasm collections in the world Country Global Institution International Crop Research Institute for the Semi-Arid Tropics (ICRISAT), Patancheru, India USA Southern Regional Plant Introduction Station, University of Georgia National Center for Genetic Resources Preservation (NCGRP) China Institute of Crop Science, Chinese Academy of Agricultural Sciences (ICS-CAAS) India National Bureau of Plant Genetic Resources (NBPGR), New Delhi Ethiopia Institute of Biodiversity Conservation (IBC) Brazil Embrapa Milho e Sorgo (CNPMS) World total Source: http: //apps 3. fao. org/wiews accessed on 29 Feb 2016 Wild 461 Cultivated Total holding (%) 39, 462 39, 923 (16. 8) 197 35, 976 36, 173 (15. 3) 2 7, 535 7, 537 (3. 2) 18, 263 (7. 7) 2, 674 14, 792 17, 466 (7. 4) 9, 772 (4. 1) 4, 017 7, 225 2, 32, 970 7, 225 (3. 0) 2, 36, 987

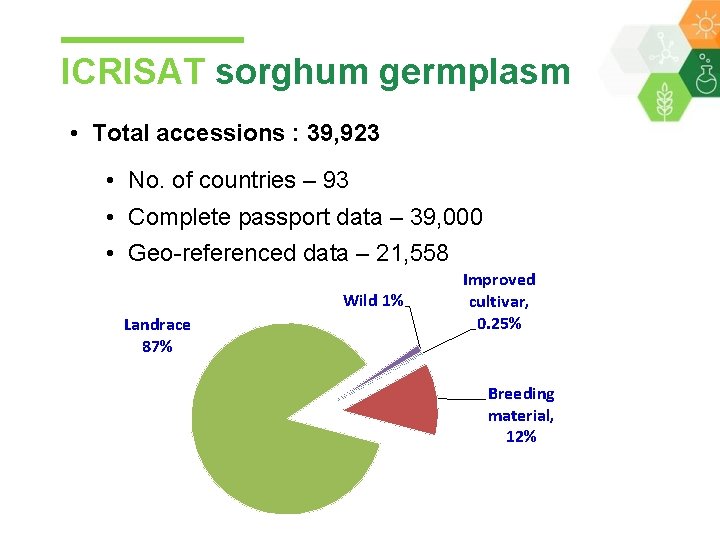
ICRISAT sorghum germplasm • Total accessions : 39, 923 • No. of countries – 93 • Complete passport data – 39, 000 • Geo-referenced data – 21, 558 Wild 1% Landrace 87% Improved cultivar, 0. 25% Breeding material, 12%

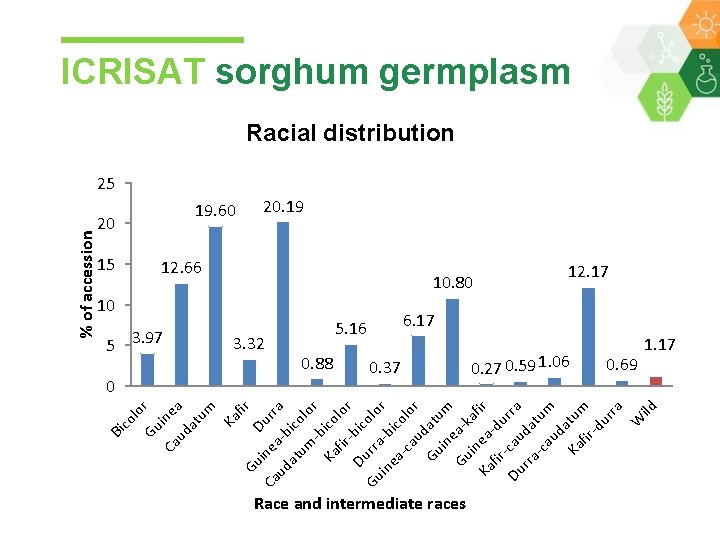
lo Gu r Ca ine ud a at um Ka fir Gu D Ca ine urr ud a-b a i at um colo -b r Ka ico fir lor Du -bic Gu rra olo r in ea bic -c olo au r Gu datu in m e a Gu in kafi r Ka ea fir -du Du -cau rra da -c tum au da Ka tum fir -d ur ra W ild Bi co % of accession ICRISAT sorghum germplasm Racial distribution 25 20 19. 60 15 5 3. 97 0 20. 19 12. 66 10. 80 10 3. 32 5. 16 0. 88 0. 37 Race and intermediate races 12. 17 6. 17 0. 27 0. 59 1. 06 0. 69 1. 17

Safety duplication of sorghum • 36, 271 accessions at Global Seed Vault at Svalbard, Norway

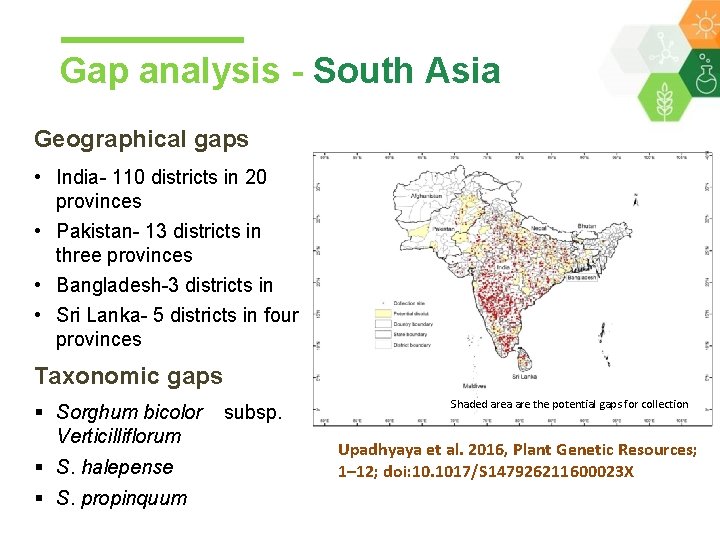
Gap analysis - South Asia Geographical gaps • India- 110 districts in 20 provinces • Pakistan- 13 districts in three provinces • Bangladesh-3 districts in • Sri Lanka- 5 districts in four provinces Taxonomic gaps § Sorghum bicolor Verticilliflorum § S. halepense § S. propinquum subsp. Shaded area are the potential gaps for collection Upadhyaya et al. 2016, Plant Genetic Resources; 1– 12; doi: 10. 1017/S 147926211600023 X

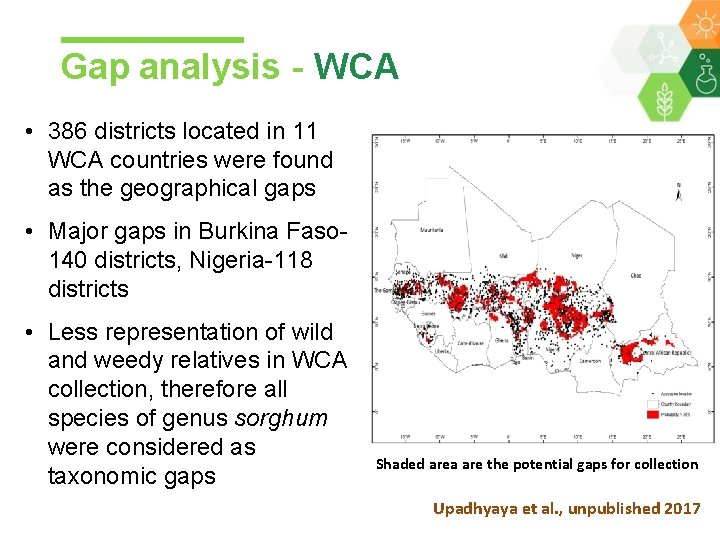
Gap analysis - WCA • 386 districts located in 11 WCA countries were found as the geographical gaps • Major gaps in Burkina Faso 140 districts, Nigeria-118 districts • Less representation of wild and weedy relatives in WCA collection, therefore all species of genus sorghum were considered as taxonomic gaps Shaded area are the potential gaps for collection Upadhyaya et al. , unpublished 2017

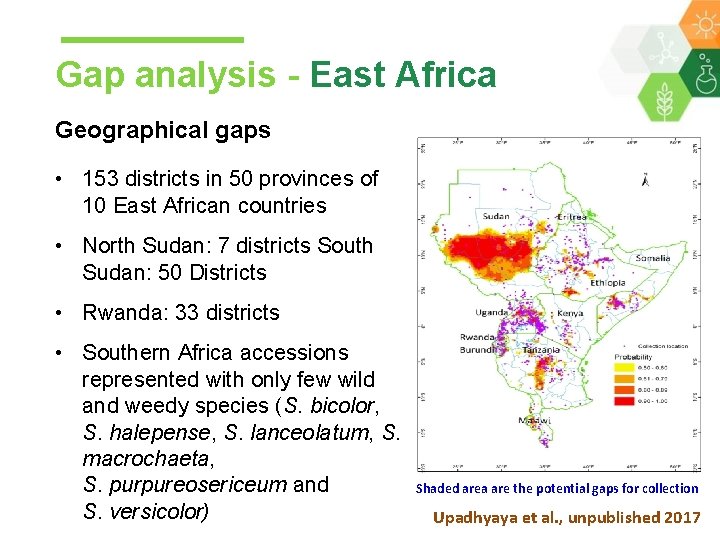
Gap analysis - East Africa Geographical gaps • 153 districts in 50 provinces of 10 East African countries • North Sudan: 7 districts South Sudan: 50 Districts • Rwanda: 33 districts • Southern Africa accessions represented with only few wild and weedy species (S. bicolor, S. halepense, S. lanceolatum, S. macrochaeta, S. purpureosericeum and S. versicolor) Shaded area are the potential gaps for collection Upadhyaya et al. , unpublished 2017

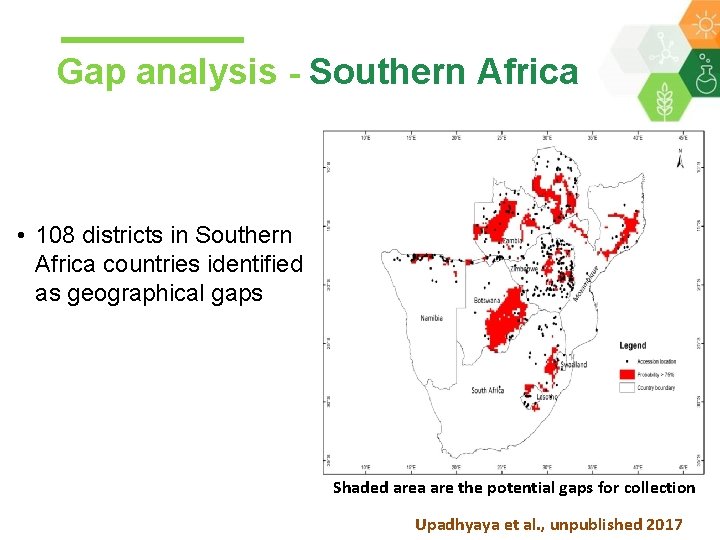
Gap analysis - Southern Africa • 108 districts in Southern Africa countries identified as geographical gaps Shaded area are the potential gaps for collection Upadhyaya et al. , unpublished 2017

Sorghum germplasm representative subsets

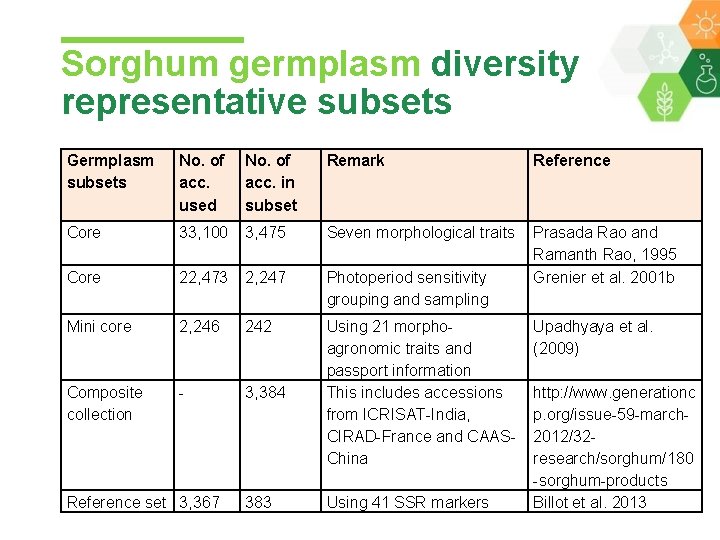
Sorghum germplasm diversity representative subsets Germplasm subsets No. of acc. used No. of acc. in subset Remark Reference Core 33, 100 3, 475 Seven morphological traits Core 22, 473 2, 247 Photoperiod sensitivity grouping and sampling Prasada Rao and Ramanth Rao, 1995 Grenier et al. 2001 b Mini core 2, 246 242 Composite collection - 3, 384 Using 21 morphoagronomic traits and passport information This includes accessions from ICRISAT-India, CIRAD-France and CAASChina Using 41 SSR markers Reference set 3, 367 383 Upadhyaya et al. (2009) http: //www. generationc p. org/issue-59 -march 2012/32 research/sorghum/180 -sorghum-products Billot et al. 2013

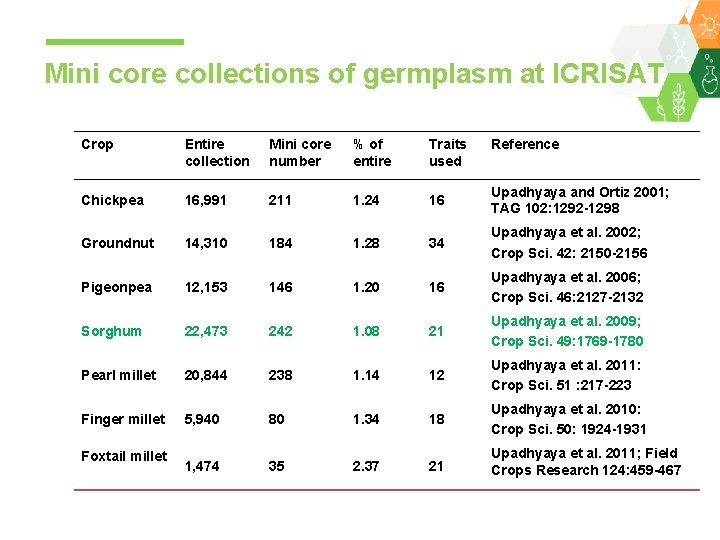
Mini core collections of germplasm at ICRISAT Crop Entire collection Mini core number % of entire Traits used Chickpea 16, 991 211 1. 24 16 Upadhyaya and Ortiz 2001; TAG 102: 1292 -1298 Groundnut 14, 310 184 1. 28 34 Upadhyaya et al. 2002; Crop Sci. 42: 2150 -2156 Pigeonpea 12, 153 146 1. 20 16 Upadhyaya et al. 2006; Crop Sci. 46: 2127 -2132 Sorghum 22, 473 242 1. 08 21 Upadhyaya et al. 2009; Crop Sci. 49: 1769 -1780 Pearl millet 20, 844 238 1. 14 12 Upadhyaya et al. 2011: Crop Sci. 51 : 217 -223 Finger millet 5, 940 80 1. 34 18 Upadhyaya et al. 2010: Crop Sci. 50: 1924 -1931 1, 474 35 2. 37 21 Upadhyaya et al. 2011; Field Crops Research 124: 459 -467 Foxtail millet Reference

Sorghum Mini Core as a Source for Traits of Economic Importance for Sorghum Improvement

Sorghum mini core collection as a sources for abiotic stress tolerance Postflowering drought and low temperature stress tolerance § 7 accessions for postflowering drought stress § 6 accessions for seedling vigor under low temperature stress § 5 accession for germinability under low temperature Postflowering drought tolerance IS# 14779, 23891, 31714, 4515, 5094, 9108, 15466 Upadhyaya et al. 2017. Crop Sci. doi: 10. 2135/cropsci 2016. 04. 0280 Seedling vigor under low temperature stress IS# 1212, 14779, 15170, 22986, 7305, 7310 Upadhyaya et al. 2016; Genome 59: 137 -145 Germinability under low temperature stress IS# 602, 1233, 7305, 10302, 20956 Upadhyaya et al. 2016; Genome 59: 137 -145

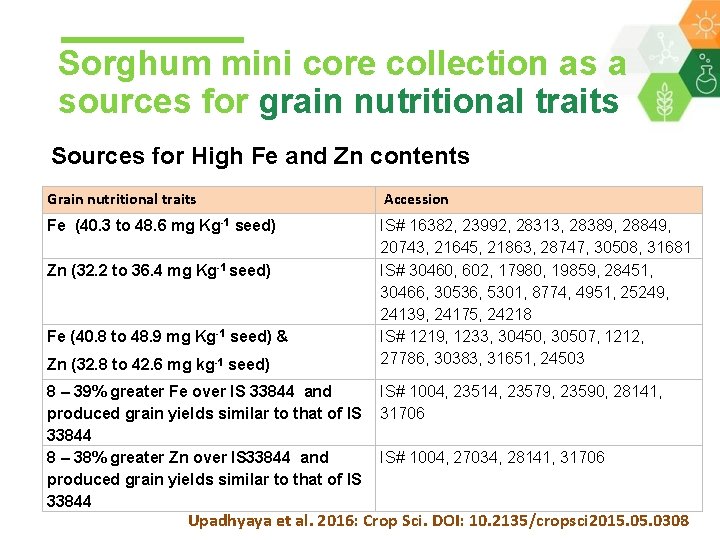
Sorghum mini core collection as a sources for grain nutritional traits Sources for High Fe and Zn contents Grain nutritional traits Accession Fe (40. 3 to 48. 6 mg Kg-1 seed) IS# 16382, 23992, 28313, 28389, 28849, 20743, 21645, 21863, 28747, 30508, 31681 IS# 30460, 602, 17980, 19859, 28451, 30466, 30536, 5301, 8774, 4951, 25249, 24139, 24175, 24218 IS# 1219, 1233, 30450, 30507, 1212, 27786, 30383, 31651, 24503 Zn (32. 2 to 36. 4 mg Kg-1 seed) Fe (40. 8 to 48. 9 mg Kg-1 seed) & Zn (32. 8 to 42. 6 mg kg-1 seed) 8 – 39% greater Fe over IS 33844 and IS# 1004, 23514, 23579, 23590, 28141, produced grain yields similar to that of IS 31706 33844 8 – 38% greater Zn over IS 33844 and IS# 1004, 27034, 28141, 31706 produced grain yields similar to that of IS 33844 Upadhyaya et al. 2016: Crop Sci. DOI: 10. 2135/cropsci 2015. 0308

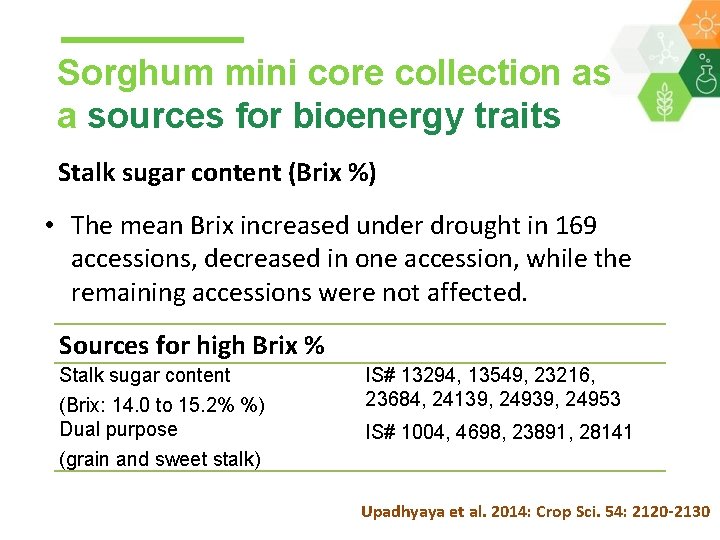
Sorghum mini core collection as a sources for bioenergy traits Stalk sugar content (Brix %) • The mean Brix increased under drought in 169 accessions, decreased in one accession, while the remaining accessions were not affected. Sources for high Brix % Stalk sugar content (Brix: 14. 0 to 15. 2% %) Dual purpose (grain and sweet stalk) IS# 13294, 13549, 23216, 23684, 24139, 24953 IS# 1004, 4698, 23891, 28141 Upadhyaya et al. 2014: Crop Sci. 54: 2120 -2130

Sorghum mini core collection as a sources for biotic stress tolerance Grain mold and Downy mildew • Grain mold resistant- 50 • Downy mildew resistant- 6 • IS 23992 resistant to both grain mold and downy mildew Downy mildew resistance IS# 28747, 31714, 23992, 27697, 28449, 30400 Grain Mold Resistance IS# 602, 603, 608, 1233, 2413, 3121, 12697, 12804, 20727, 20740, 20743, 20816, 30562, 31681, 2379, 2864, 12302, 13971, 17941, 19389, 23992, 26694, 29335, 21512, 21645, 12945, 22294, 995, 2426, 12706, 16151, 24453, 26701, 29326, 30383, 30536, 20956, 29314, 30092, 10969, 23590, 29187, 29269, 473, 29304, 1212, 13893, 29241, 29568 Sharma et al. , 2010 (Plant Disease 94: 439 -444)

Sorghum mini core collection as a sources for biotic stress tolerance Foliar diseases • 13 accessions resistance to anthracnose, 27 resistant to leaf blight and 6 resistant to rust identified Anthracnose resistance IS# 473, 5301, 6354, 7679, 10302, 16382, 19153, Sharma et al. 2012 (Plant 20632, 20956, 23521, 23684, 24218, 24939 Disease 96: 1629 -1633) Leaf blight resistance IS# 473, 2382, 7131, 9108, 9177, 9745, 12937, 12945, 14861, 19445, 20743, 21083, 23521, 23644, 23684, 24175, 24503, 24939, 24953, 26694, 26749, 28614, 29187, 29233, 29714, 31557, 33353 Rust resistance IS# 473, 23521, 23684, 24503, 26737, 33023 Sharma et al. , 2012 (Plant Disease 96: 1629 -1633)

Sorghum mini core collection as a sources for biotic stress tolerance Resistance to multiple diseases • IS 473 for grain mold, anthracnose, leaf blight and rust • IS 23684 and IS 23521 for anthracnose, leaf blight and rust; • IS 24939 for anthracnose and leaf blight • IS 23992 for grain molds and downy mildew • IS 12945, IS 26694, IS 29187 for grain mold and leaf blight; • IS 20956 for grain mold anthracnose The accessions with multiple disease resistance will be useful in sorghum disease resistance breeding programs

Sorghum Genomic Resources

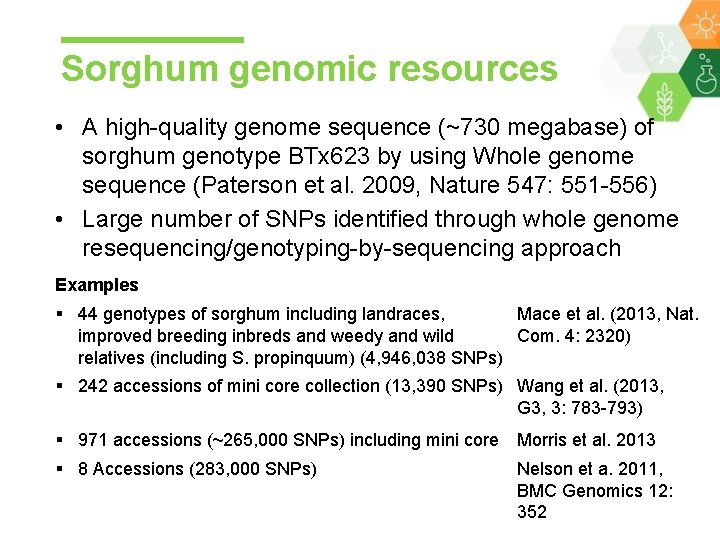
Sorghum genomic resources • A high-quality genome sequence (~730 megabase) of sorghum genotype BTx 623 by using Whole genome sequence (Paterson et al. 2009, Nature 547: 551 -556) • Large number of SNPs identified through whole genome resequencing/genotyping-by-sequencing approach Examples § 44 genotypes of sorghum including landraces, Mace et al. (2013, Nat. improved breeding inbreds and weedy and wild Com. 4: 2320) relatives (including S. propinquum) (4, 946, 038 SNPs) § 242 accessions of mini core collection (13, 390 SNPs) Wang et al. (2013, G 3, 3: 783 -793) § 971 accessions (~265, 000 SNPs) including mini core Morris et al. 2013 § 8 Accessions (283, 000 SNPs) Nelson et a. 2011, BMC Genomics 12: 352

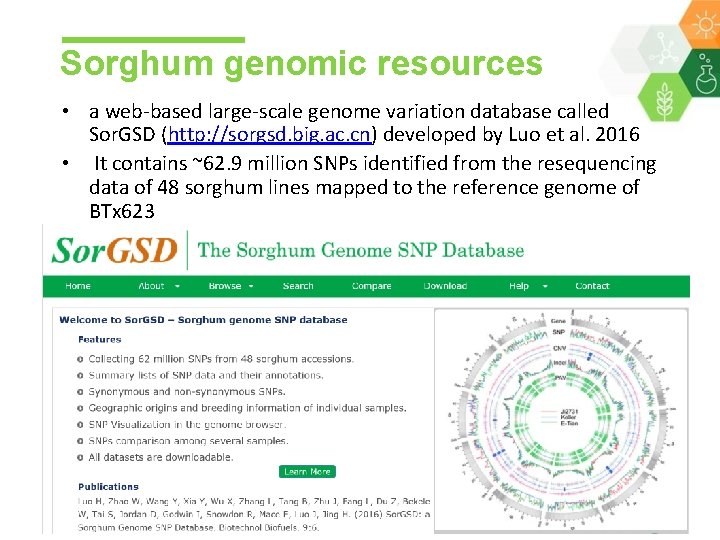
Sorghum genomic resources • a web-based large-scale genome variation database called Sor. GSD (http: //sorgsd. big. ac. cn) developed by Luo et al. 2016 • It contains ~62. 9 million SNPs identified from the resequencing data of 48 sorghum lines mapped to the reference genome of BTx 623

Genotyping germplasm subsets: Population structure and diversity

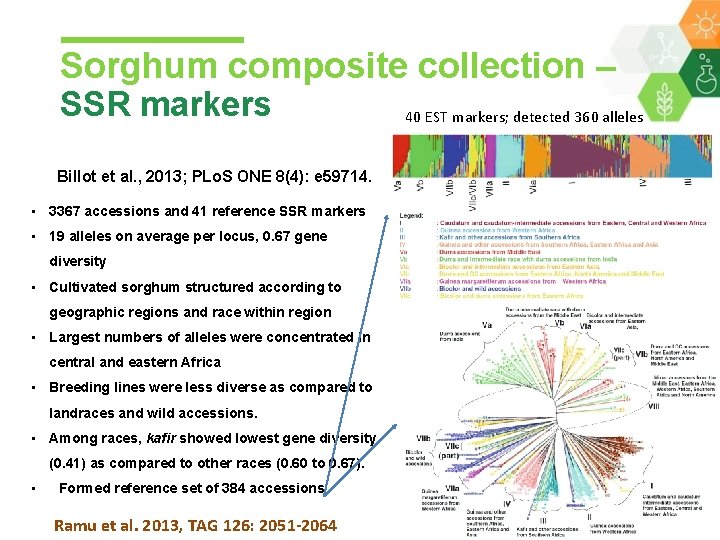
Sorghum composite collection – SSR markers Billot et al. , 2013; PLo. S ONE 8(4): e 59714. • 3367 accessions and 41 reference SSR markers • 19 alleles on average per locus, 0. 67 gene diversity • Cultivated sorghum structured according to geographic regions and race within region • Largest numbers of alleles were concentrated in central and eastern Africa • Breeding lines were less diverse as compared to landraces and wild accessions. • Among races, kafir showed lowest gene diversity (0. 41) as compared to other races (0. 60 to 0. 67). • Formed reference set of 384 accessions Ramu et al. 2013, TAG 126: 2051 -2064 40 EST markers; detected 360 alleles

Genotyping-by-Sequencing (GBS) of large sorghum collection • Diverse sorghum germplasm (971 accessions) from worldwide collections by GBS approach (∼ 265, 000 SNPs) • 971 accessions including. . – the US association panel (Casa et al. , 2008), – the mini core collection (Upadhyaya et al. , 2009) and – reference set (Billot et al. , 2013)

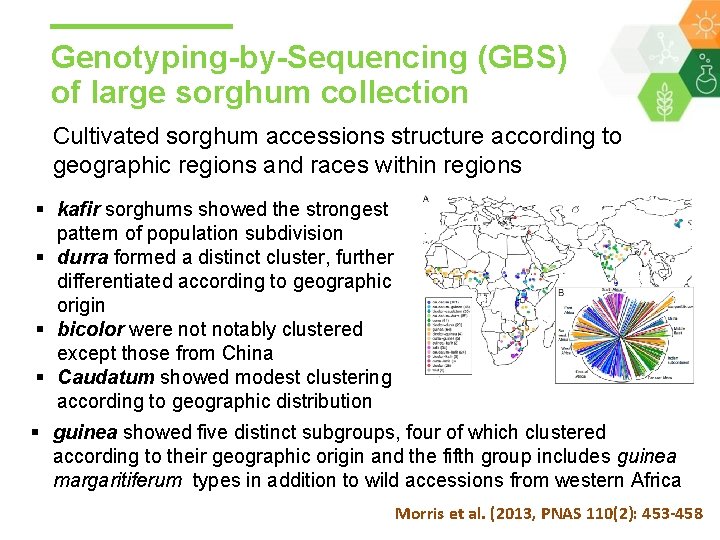
Genotyping-by-Sequencing (GBS) of large sorghum collection Cultivated sorghum accessions structure according to geographic regions and races within regions § kafir sorghums showed the strongest pattern of population subdivision § durra formed a distinct cluster, further differentiated according to geographic origin § bicolor were notably clustered except those from China § Caudatum showed modest clustering according to geographic distribution § guinea showed five distinct subgroups, four of which clustered according to their geographic origin and the fifth group includes guinea margaritiferum types in addition to wild accessions from western Africa Morris et al. (2013, PNAS 110(2): 453 -458

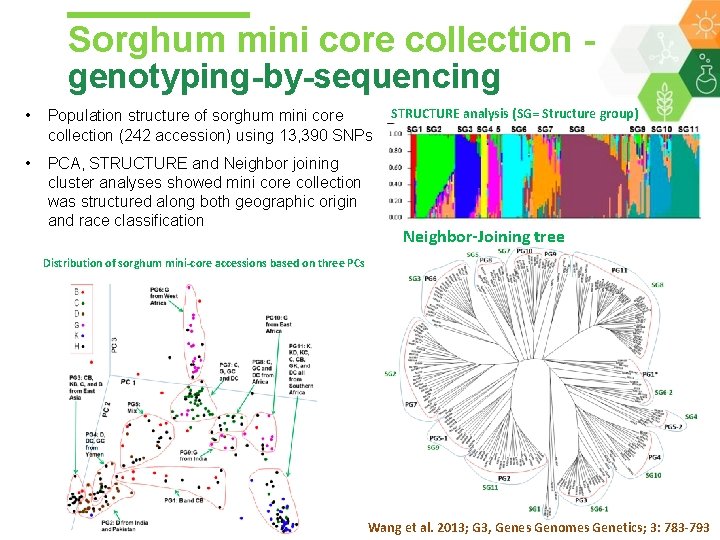
Sorghum mini core collection - genotyping-by-sequencing • Population structure of sorghum mini core collection (242 accession) using 13, 390 SNPs • PCA, STRUCTURE and Neighbor joining cluster analyses showed mini core collection was structured along both geographic origin and race classification STRUCTURE analysis (SG= Structure group) Neighbor-Joining tree Distribution of sorghum mini-core accessions based on three PCs Wang et al. 2013; G 3, Genes Genomes Genetics; 3: 783 -793

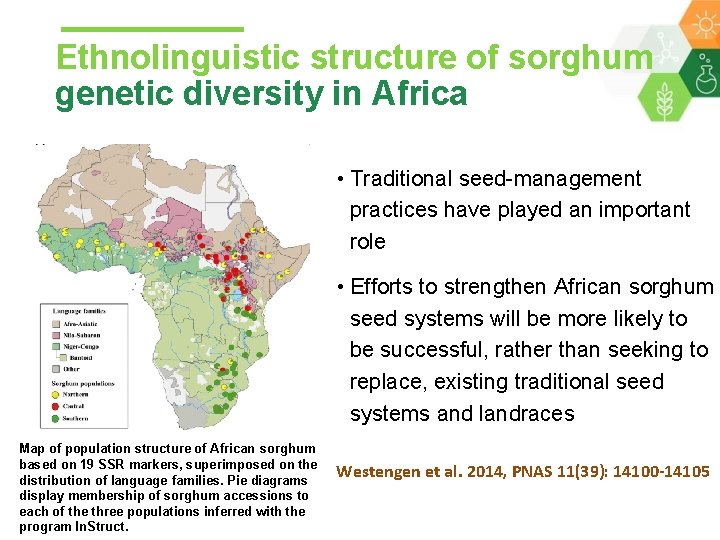
Ethnolinguistic structure of sorghum genetic diversity in Africa • Cultural factors play a key role in shaping the genetic structure in sorghum • Close associations between sorghum population structure and the distribution of ethnolinguistic groups in Africa

Ethnolinguistic structure of sorghum genetic diversity in Africa • Traditional seed-management practices have played an important role • Efforts to strengthen African sorghum seed systems will be more likely to be successful, rather than seeking to replace, existing traditional seed systems and landraces Map of population structure of African sorghum based on 19 SSR markers, superimposed on the distribution of language families. Pie diagrams display membership of sorghum accessions to each of the three populations inferred with the program In. Struct. Westengen et al. 2014, PNAS 11(39): 14100 -14105

Genome-wide Association Studies

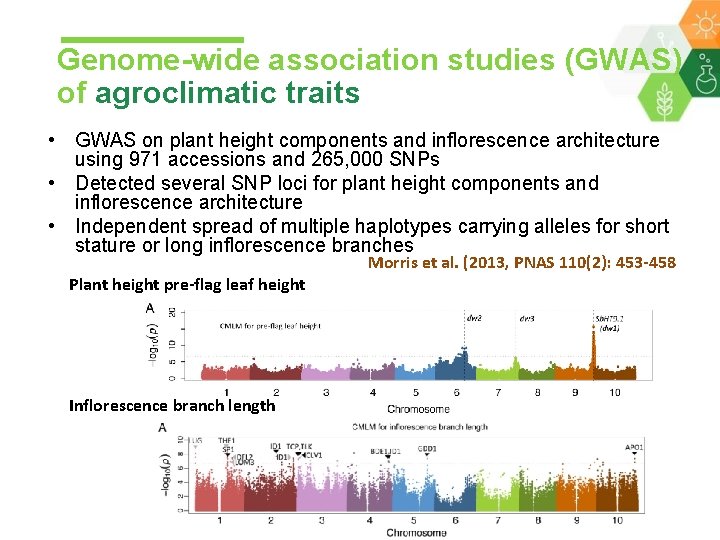
Genome-wide association studies (GWAS) of agroclimatic traits • GWAS on plant height components and inflorescence architecture using 971 accessions and 265, 000 SNPs • Detected several SNP loci for plant height components and inflorescence architecture • Independent spread of multiple haplotypes carrying alleles for short stature or long inflorescence branches Plant height pre-flag leaf height Inflorescence branch length Morris et al. (2013, PNAS 110(2): 453 -458

Genome-environment associations in sorghum landraces predict adaptive traits Ho: If association between SNP variation and environ. of origin in landraces reflect adaptation, then these could predict adaptive traits. -1, 943 georeferenced landraces, 404, 627 SNPs. • Environment explained a substantial portion of SNP variation • Environment-associated SNPs predicted genotype-by-environment interactions under experimental drought stress and aluminum toxicity • Genomic signatures of environmental adaptation useful for crop improvement, enhancing germplasm identification • Together, genome-environment associations and phenotypic analyses revealed the basis of environmental adaptation

Genome-environment associations in sorghum landraces predict adaptive traits Testing Ho: used mini core post-flowering drought at ICRISAT and Austin and Al toxicity at Austin • Change in panicle seed weight from well-watered (WW) to drought (D) best predicted by SNPs with strongest association with season length - Accessions with alleles associated with long growing season had larger reduction compared to accessions with alleles associated with short growing season - alleles selected for in short growing season may confer drought tolerance • Change in harvest index (HI) from WW to D in Austin was best predicted by SNP with strongest association with precipitation in warmest quarter - Accessions with alleles associated with wet warmest quarters showed large reductions in HI from WW to D - Accessions with alleles associated with dry warmest quarters often showed high HI in D than in WW

Sorghum mini core collection as an association mapping panel • Sorghum mini core collection used as an association mapping panel for genetic dissection of complex traits mainly because of - Manageable size (242 accessions) - Representing diversity of entire collection - Included all races and intermediate races - Diverse origin (57 countries from Africa, the Americas, Asia, Mediterranean, Oceania)

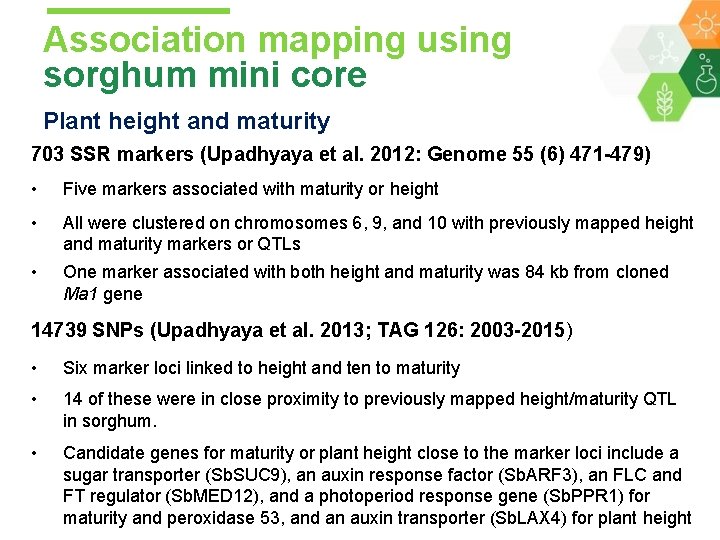
Association mapping using sorghum mini core Plant height and maturity 703 SSR markers (Upadhyaya et al. 2012: Genome 55 (6) 471 -479) • Five markers associated with maturity or height • All were clustered on chromosomes 6, 9, and 10 with previously mapped height and maturity markers or QTLs • One marker associated with both height and maturity was 84 kb from cloned Ma 1 gene 14739 SNPs (Upadhyaya et al. 2013; TAG 126: 2003 -2015) • Six marker loci linked to height and ten to maturity • 14 of these were in close proximity to previously mapped height/maturity QTL in sorghum. • Candidate genes for maturity or plant height close to the marker loci include a sugar transporter (Sb. SUC 9), an auxin response factor (Sb. ARF 3), an FLC and FT regulator (Sb. MED 12), and a photoperiod response gene (Sb. PPR 1) for maturity and peroxidase 53, and an auxin transporter (Sb. LAX 4) for plant height

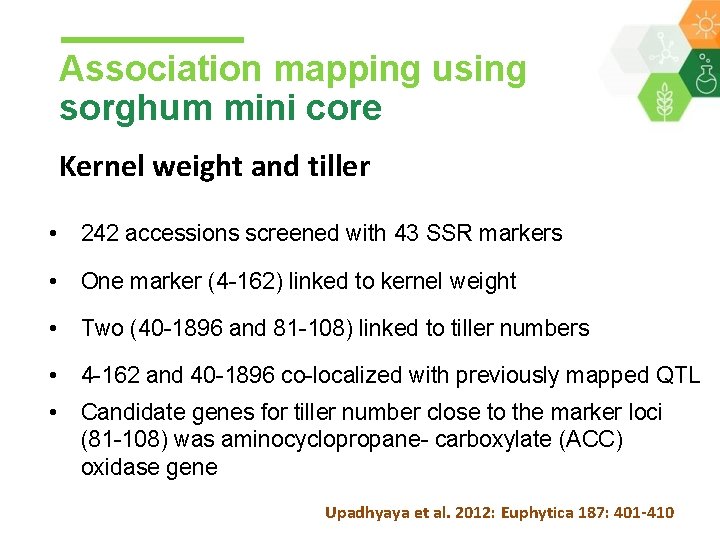
Association mapping using sorghum mini core Kernel weight and tiller • 242 accessions screened with 43 SSR markers • One marker (4 -162) linked to kernel weight • Two (40 -1896 and 81 -108) linked to tiller numbers • 4 -162 and 40 -1896 co-localized with previously mapped QTL • Candidate genes for tiller number close to the marker loci (81 -108) was aminocyclopropane- carboxylate (ACC) oxidase gene Upadhyaya et al. 2012: Euphytica 187: 401 -410

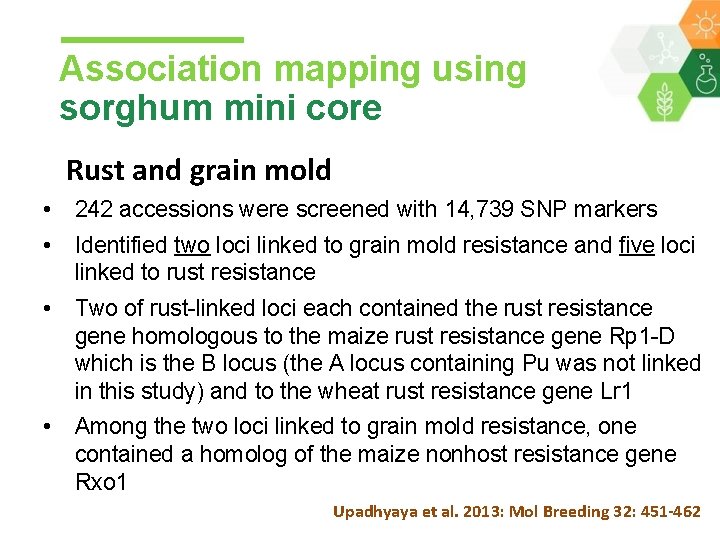
Association mapping using sorghum mini core Rust and grain mold • 242 accessions were screened with 14, 739 SNP markers • Identified two loci linked to grain mold resistance and five loci linked to rust resistance • Two of rust-linked loci each contained the rust resistance gene homologous to the maize rust resistance gene Rp 1 -D which is the B locus (the A locus containing Pu was not linked in this study) and to the wheat rust resistance gene Lr 1 • Among the two loci linked to grain mold resistance, one contained a homolog of the maize nonhost resistance gene Rxo 1 Upadhyaya et al. 2013: Mol Breeding 32: 451 -462

Association mapping using sorghum mini core Anthracnose resistance • 242 accessions were screened with 14, 739 SNP markers • Identified 8 marker loci (loci 1 -8) were associated with anthracnose resistance • These include two NB-ARC class of R gene on chromosome 10 that were partially homologous to the rice blast gene Pib

Association mapping using sorghum mini core Germination under low temperature § Early-season planting extends sorghum’s growing season and increases yield in temperate regions § However, sorghum’s sensitivity to low soil temperatures adversely impacts seed germination § GWAS using 162, 177 SNPs identified one marker locus (Locus 7 -2) as significantly associated with low-temperature germination. Identified SNP co-localized • With previously mapped cold tolerance quantitative trait loci (QTL) in sorghum, • With a candidate gene that increases cold tolerance and germination rate when its wheat homolog is overexpressed in tobacco, and • Its syntenic region in rice co-localized with two cold tolerance QTL in rice Upadhyaya et al. 2016: Genome 59: 137 -145

Association mapping using sorghum mini core Saccharification yield • 242 accessions of sorghum mini core germplasm collection and 14, 739 SNPs • Identified 7 SNP loci associated with Saccharification yield and five of these loci were syntenic with regions in the maize genome that contain quantitative trait loci underlying saccharification yield and cell wall component traits • The most promising candidates are – β- tubulin that determines the orientation of cellulose microfibrils in plant secondary cell walls, and – NST 1, a master transcription factor controlling secondary cell wall biosynthesis in fibers Wang et al. 2013, Genome 56: 659 -665

Conclusions-Enhancing use of germpalsm • Due to reduced size and representativeness of species diversity, the mini core collections are an ideal genetic resources for an in depth characterization of its biological diversity and use in crop improvement programs (Upadhyaya and Ortiz, 2001) • Mini core collections developed at ICRISAT genebank are now International Public Goods and serves as a gateway to access the species diversity. • Already provided 280 sets of mini core collections of different crops to scientists in 36 countries and 114 sets to scientists at ICRISAT

Conclusions • Mini core collection useful for identification of multi trait germplasm to meet needs of breeders for traits of economic importance and for mapping traits • Genotyping mini core helps in identifying genetically diverse parents for use in breeding and mapping but also helps in identification of duplicates, genetic drift, etc. MINI CORE WORKS

Thank You