Some new sequencing technologies Molecular Inversion Probes Illumina

Some new sequencing technologies
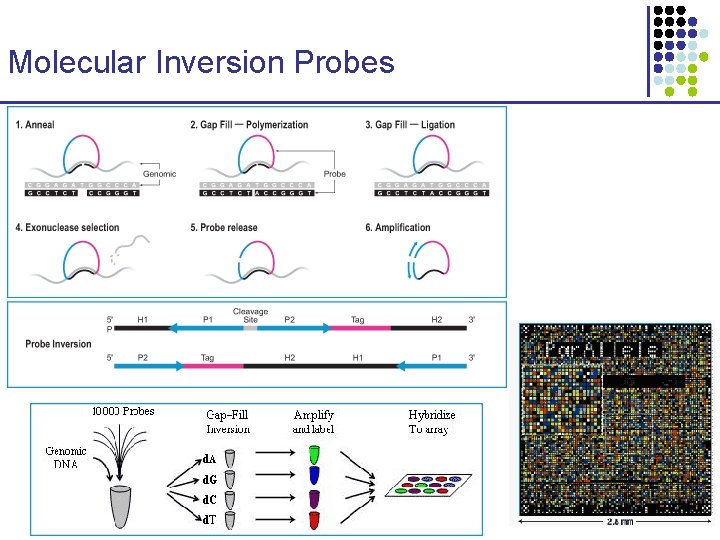
Molecular Inversion Probes
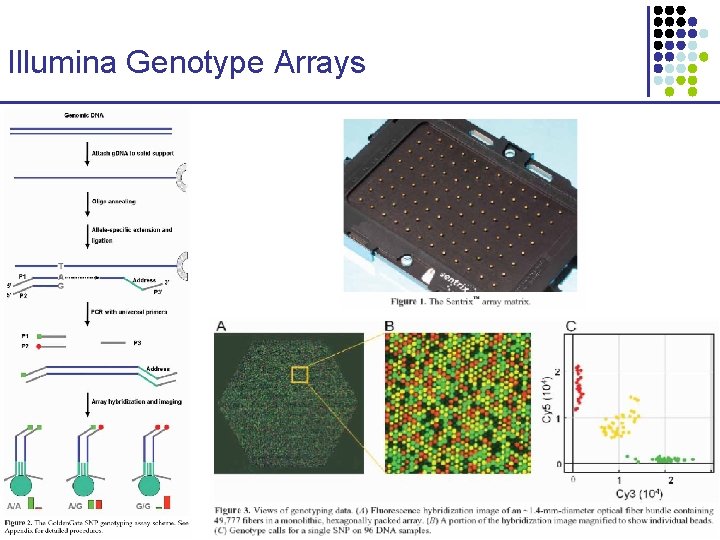
Illumina Genotype Arrays

Single Molecule Array for Genotyping—Solexa
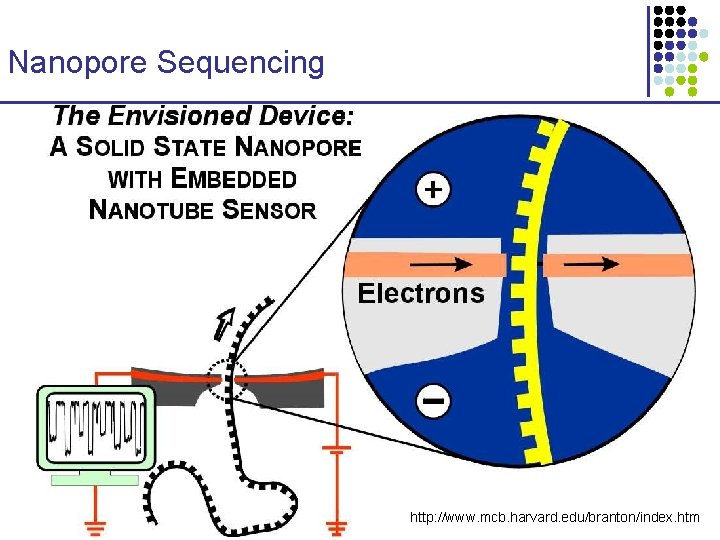
Nanopore Sequencing http: //www. mcb. harvard. edu/branton/index. htm
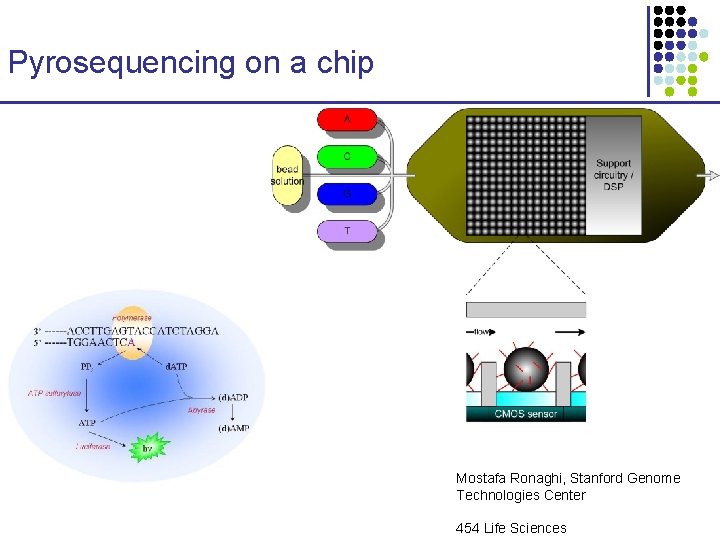
Pyrosequencing on a chip Mostafa Ronaghi, Stanford Genome Technologies Center 454 Life Sciences
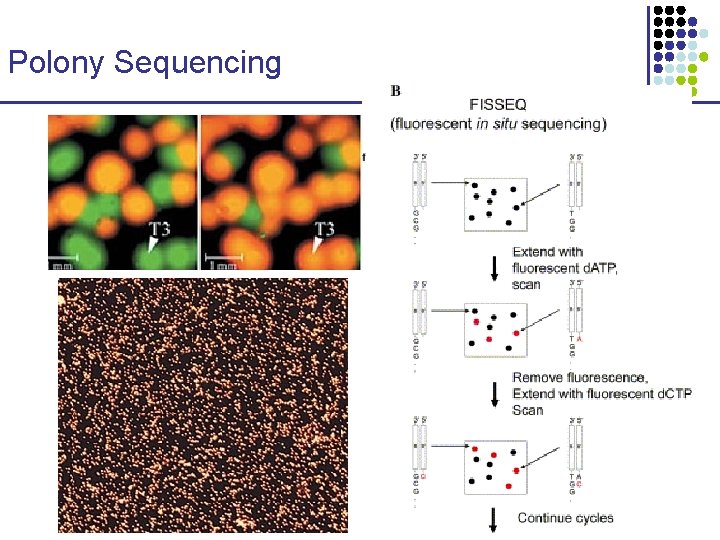
Polony Sequencing

Technologies available today • Illumina § 550, 000 SNP array: $300 -500 in bulk • 454 § 200 bp reads, 100 Mbp total sequence in 1 run, $8 K § 500 bp reads in much higher throughput coming soon • Solexa § 1 Gbp of sequence coming in paired 35 bp reads § 1 day, approx $10 K / run
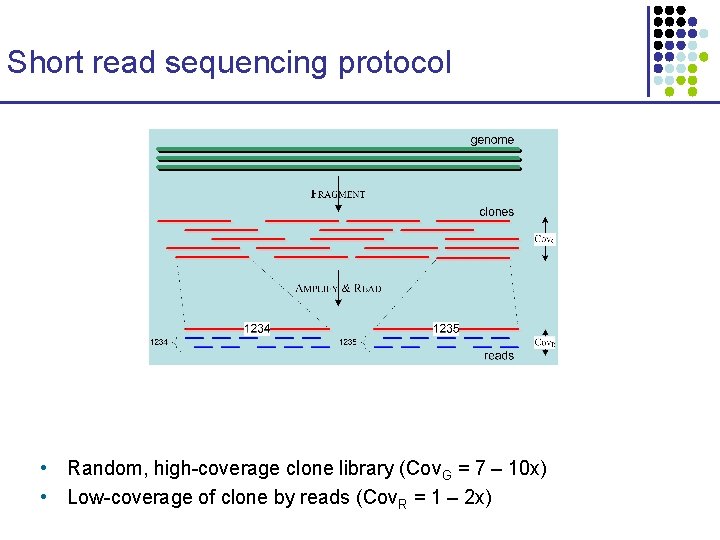
Short read sequencing protocol • Random, high-coverage clone library (Cov. G = 7 – 10 x) • Low-coverage of clone by reads (Cov. R = 1 – 2 x)
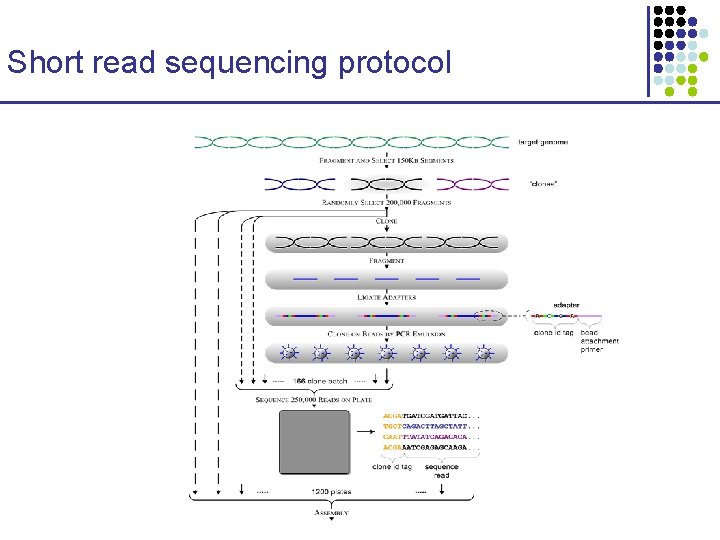
Short read sequencing protocol
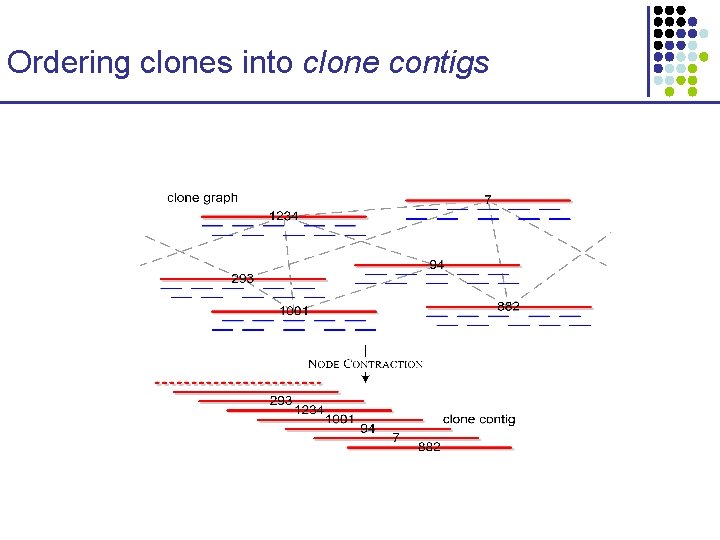
Ordering clones into clone contigs
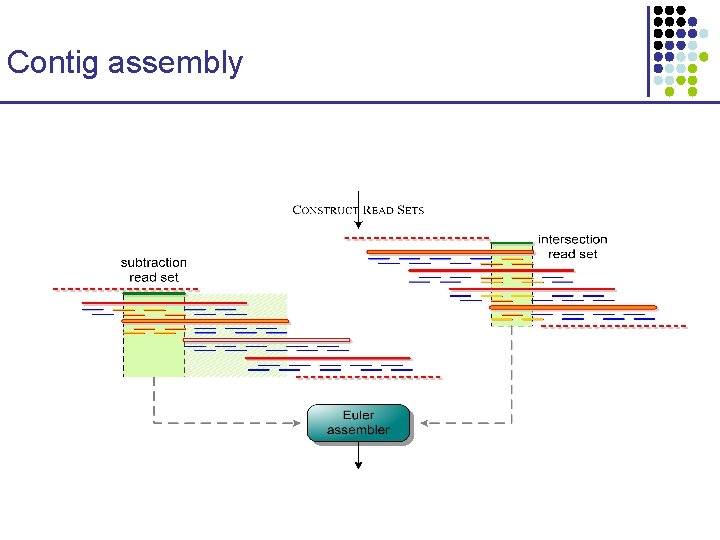
Contig assembly
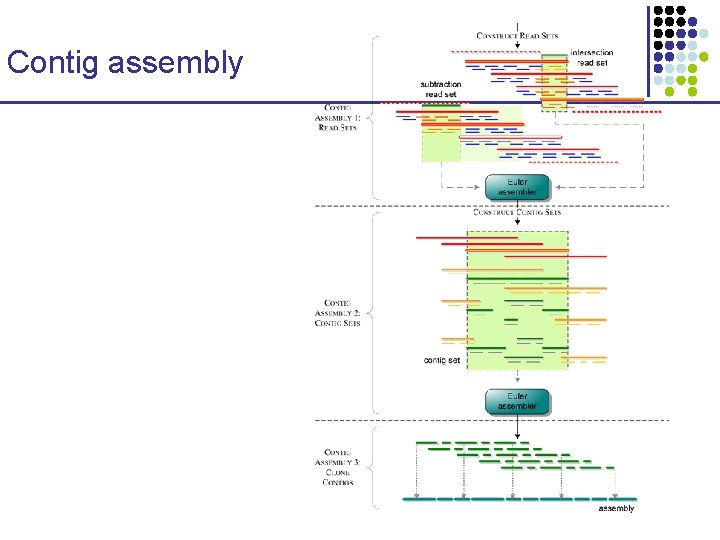
Contig assembly
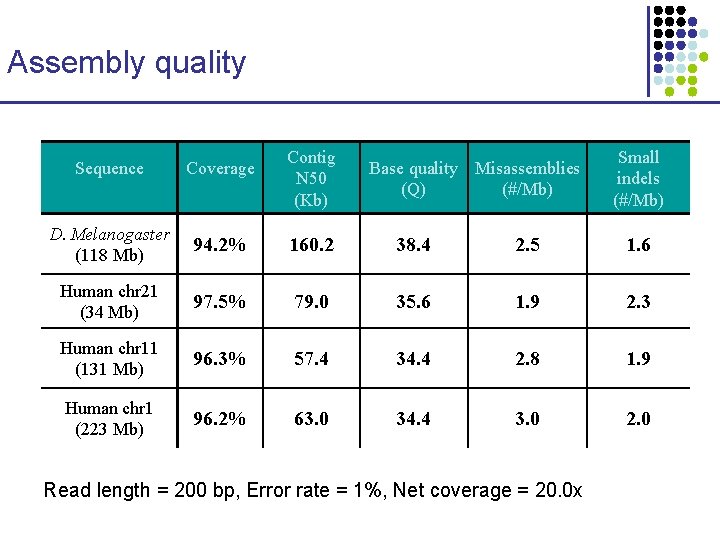
Assembly quality Contig N 50 (Kb) Base quality Misassemblies (Q) (#/Mb) Small indels (#/Mb) Sequence Coverage D. Melanogaster (118 Mb) 94. 2% 160. 2 38. 4 2. 5 1. 6 Human chr 21 (34 Mb) 97. 5% 79. 0 35. 6 1. 9 2. 3 Human chr 11 (131 Mb) 96. 3% 57. 4 34. 4 2. 8 1. 9 Human chr 1 (223 Mb) 96. 2% 63. 0 34. 4 3. 0 2. 0 Read length = 200 bp, Error rate = 1%, Net coverage = 20. 0 x

Multiple Sequence Alignment
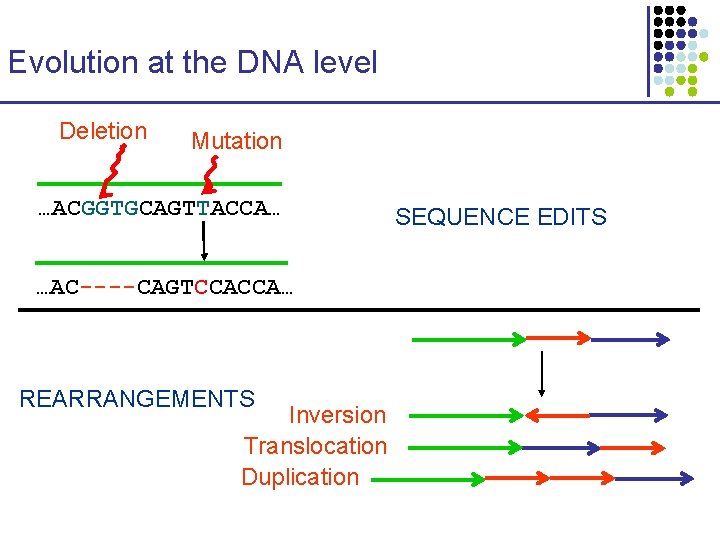
Evolution at the DNA level Deletion Mutation …ACGGTGCAGTTACCA… …AC----CAGTCCACCA… REARRANGEMENTS Inversion Translocation Duplication SEQUENCE EDITS
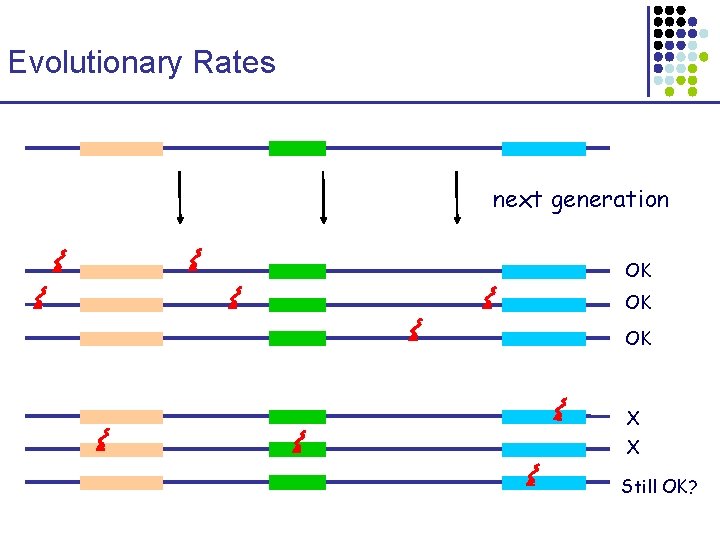
Evolutionary Rates next generation OK OK OK X X Still OK?
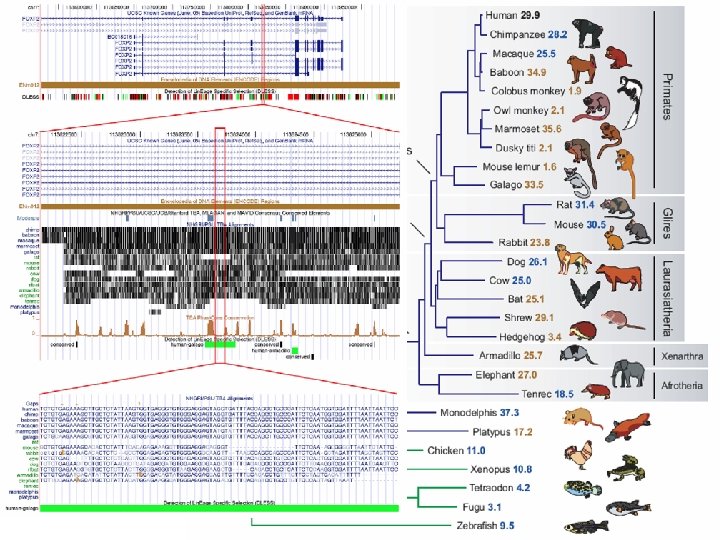
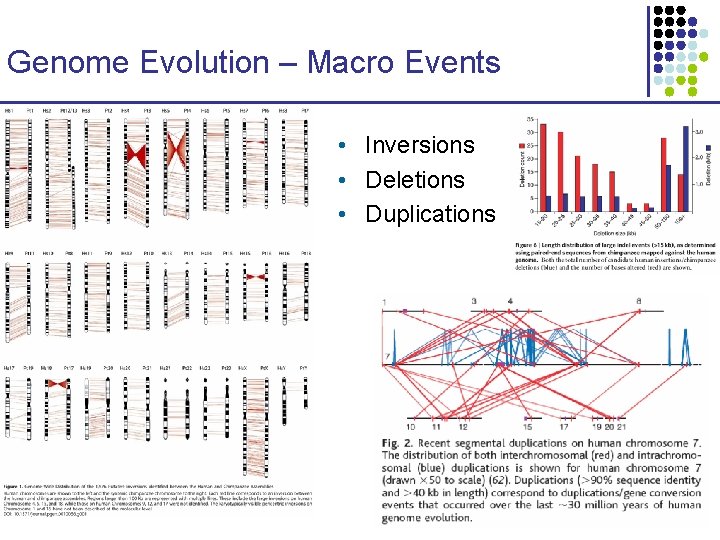
Genome Evolution – Macro Events • Inversions • Deletions • Duplications
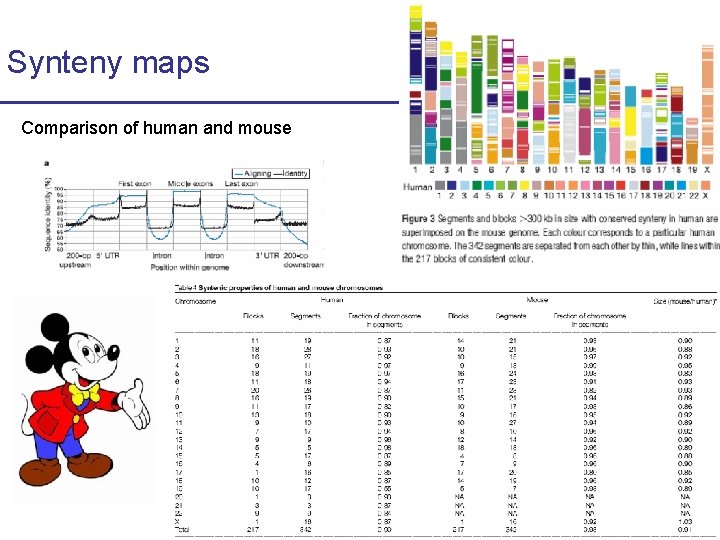
Synteny maps Comparison of human and mouse
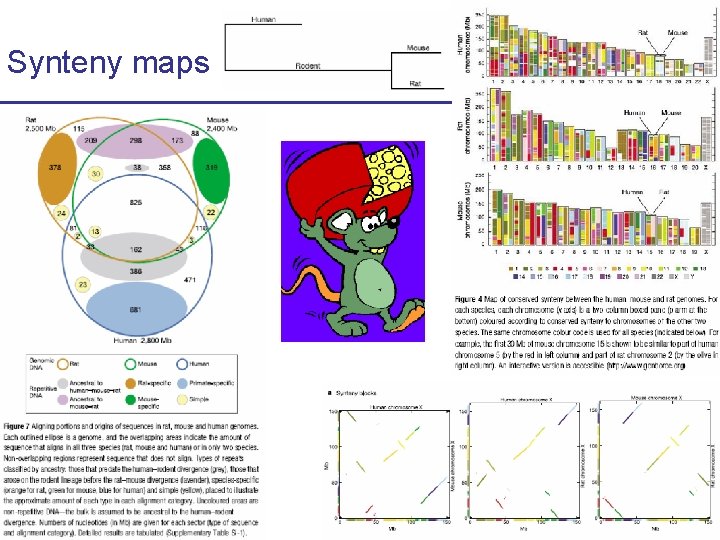
Synteny maps
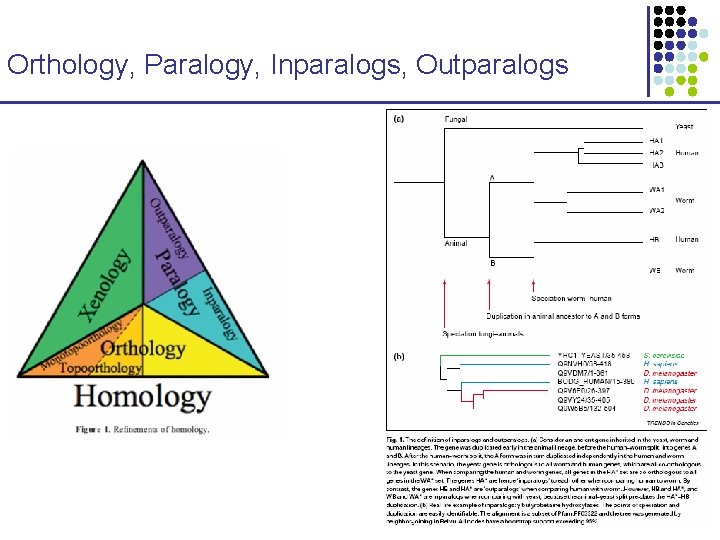
Orthology, Paralogy, Inparalogs, Outparalogs
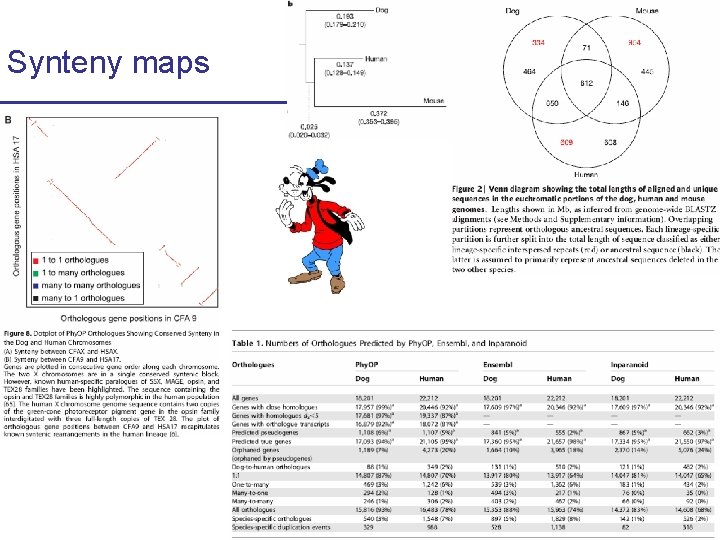
Synteny maps
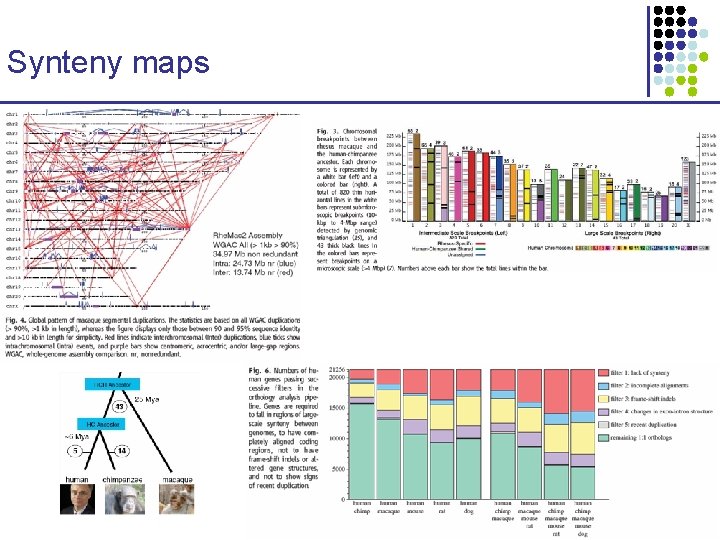
Synteny maps
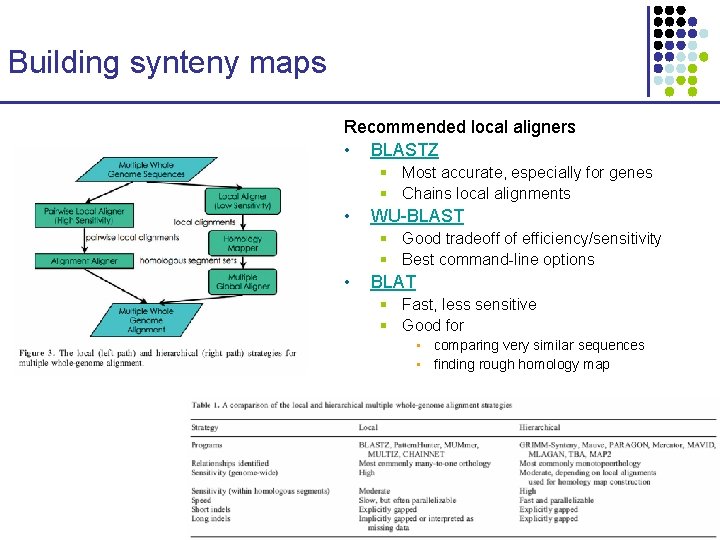
Building synteny maps Recommended local aligners • BLASTZ § Most accurate, especially for genes § Chains local alignments • WU-BLAST § Good tradeoff of efficiency/sensitivity § Best command-line options • BLAT § Fast, less sensitive § Good for • comparing very similar sequences • finding rough homology map
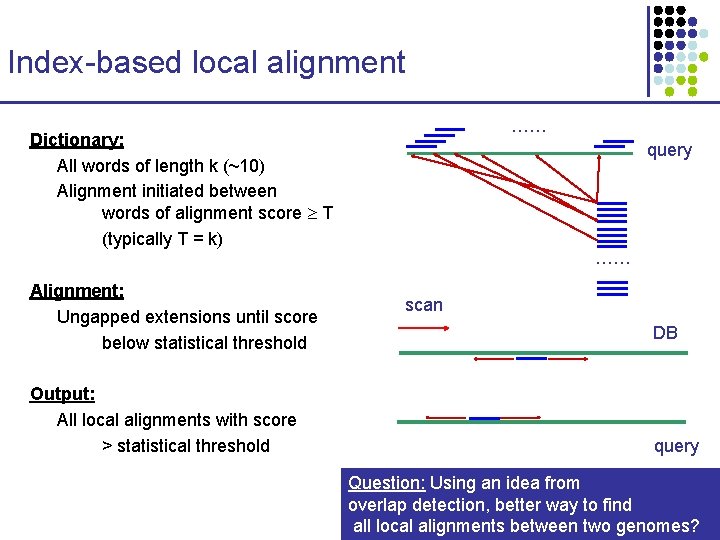
Index-based local alignment …… Dictionary: All words of length k (~10) Alignment initiated between words of alignment score T (typically T = k) Alignment: Ungapped extensions until score below statistical threshold Output: All local alignments with score > statistical threshold query …… scan DB query Question: Using an idea from overlap detection, better way to find all local alignments between two genomes?

Local Alignments
After chaining
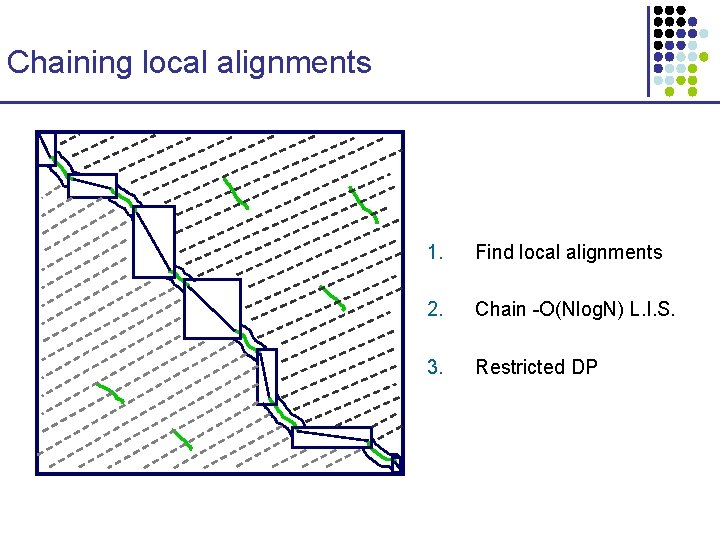
Chaining local alignments 1. Find local alignments 2. Chain -O(Nlog. N) L. I. S. 3. Restricted DP
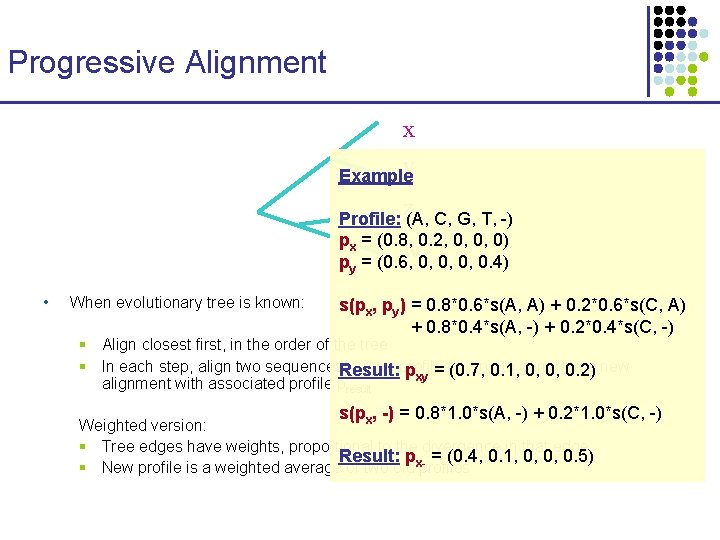
Progressive Alignment x y Example z Profile: (A, C, G, T, -) px = (0. 8, 0. 2, 0, 0, 0) w py = (0. 6, 0, 0. 4) • When evolutionary tree is known: s(px, py) = 0. 8*0. 6*s(A, A) + 0. 2*0. 6*s(C, A) + 0. 8*0. 4*s(A, -) + 0. 2*0. 4*s(C, -) § Align closest first, in the order of the tree § In each step, align two sequences. Result: x, y, or profiles px, py 0. 1, , to generate a new pxy = (0. 7, 0, 0, 0. 2) alignment with associated profile presult s(p , -) = 0. 8*1. 0*s(A, -) + 0. 2*1. 0*s(C, -) x Weighted version: § Tree edges have weights, proportional to the divergence in that edge Result: p = (0. 4, 0. 1, 0, 0, 0. 5) § New profile is a weighted average of two old x-profiles
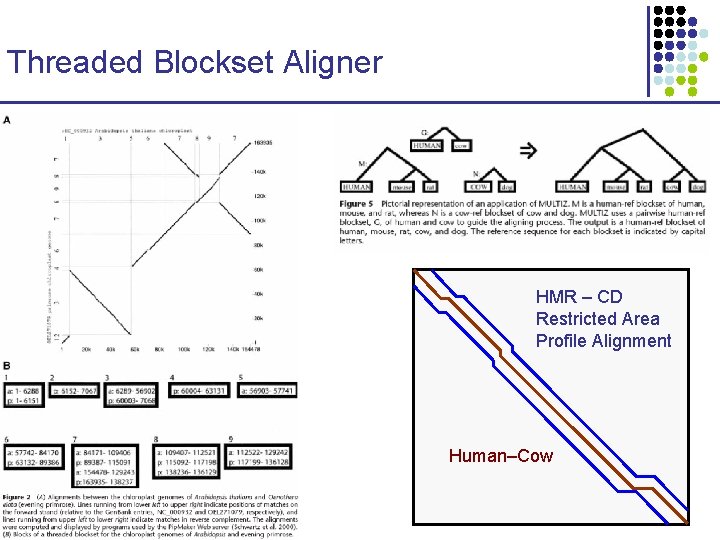
Threaded Blockset Aligner HMR – CD Restricted Area Profile Alignment Human–Cow
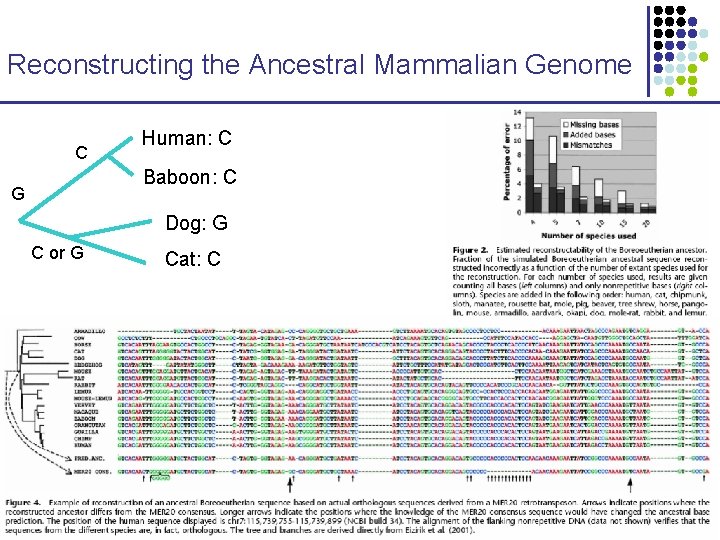
Reconstructing the Ancestral Mammalian Genome C Human: C Baboon: C G Dog: G C or G Cat: C
- Slides: 32