Software for Protein Structures by NMR Software Can
Software for Protein Structures by NMR Software Can be Grouped into Two General Classes: • Protein Based Programs: ► Calculate Protein Structures XPLOR (CNS, CXS), DYANA, CHARMM, Sybyl, Amber, etc. ► Visualize Protein Structures t. Quanta, Insight II, XPLOR-VMD, Ras. Mol, Chimera, MOLMOL, Ribbons, etc ► Evaluate Protein Structures t PROCHECK, WHATIF, Verify 3 D, etc t • NMR Based Programs ► NMR data processing NMRPipe, Felix ► NMR data analysis/visualization t NMRDraw, NMRView, PIPP ► Iterative Relaxation Matrix Calculations t IRMA, CORMA, MARDIGRAS, XPLOR, MORASS, etc ► Automated NMR Analysis t Auto. Assign, Auto. Structure, ARIA, CANDID, GARRANT, etc t t Not A complete List of Software ► New software is constantly being developed In a practical, sense, only use a small subset of available software t a lot of redundancy, use what trained on/comfortable with. ► t No Real Standards different file formats, a lot of incompatibilities and file manipulations necessary. ►
Software for Protein Structures by NMR Protein NMR Based Software Programs: • There are multiple programs that have similar functions. • Not practical or necessary to discuss all the variety of programs that are available. • Applications will be discussed in general with specific references to a limited number of programs. Protein Based Programs: Visualize Protein Structures • How is the protein structure stored? No uniformat. t Protein Data Bank (PDB) is the closest thing to a uniformed format t Most programs can read and/or write PDB file formats ► Just about every program has its own proprietary format t Babel program can interconvert ~47 different structure formats ► • Common Information in a protein structure: ► atoms, residues, chains ► X, Y, Z coordinates
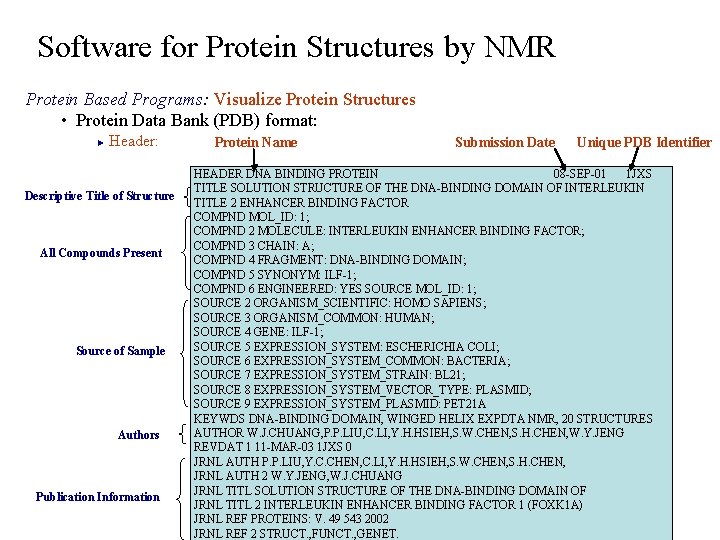
Software for Protein Structures by NMR Protein Based Programs: Visualize Protein Structures • Protein Data Bank (PDB) format: Header: ► Descriptive Title of Structure All Compounds Present Source of Sample Authors Publication Information Protein Name Submission Date Unique PDB Identifier HEADER DNA BINDING PROTEIN 08 -SEP-01 1 JXS TITLE SOLUTION STRUCTURE OF THE DNA-BINDING DOMAIN OF INTERLEUKIN TITLE 2 ENHANCER BINDING FACTOR COMPND MOL_ID: 1; COMPND 2 MOLECULE: INTERLEUKIN ENHANCER BINDING FACTOR; COMPND 3 CHAIN: A; COMPND 4 FRAGMENT: DNA-BINDING DOMAIN; COMPND 5 SYNONYM: ILF-1; COMPND 6 ENGINEERED: YES SOURCE MOL_ID: 1; SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS; SOURCE 3 ORGANISM_COMMON: HUMAN; SOURCE 4 GENE: ILF-1; SOURCE 5 EXPRESSION_SYSTEM: ESCHERICHIA COLI; SOURCE 6 EXPRESSION_SYSTEM_COMMON: BACTERIA; SOURCE 7 EXPRESSION_SYSTEM_STRAIN: BL 21; SOURCE 8 EXPRESSION_SYSTEM_VECTOR_TYPE: PLASMID; SOURCE 9 EXPRESSION_SYSTEM_PLASMID: PET 21 A KEYWDS DNA-BINDING DOMAIN, WINGED HELIX EXPDTA NMR, 20 STRUCTURES AUTHOR W. J. CHUANG, P. P. LIU, C. LI, Y. H. HSIEH, S. W. CHEN, S. H. CHEN, W. Y. JENG REVDAT 1 11 -MAR-03 1 JXS 0 JRNL AUTH P. P. LIU, Y. C. CHEN, C. LI, Y. H. HSIEH, S. W. CHEN, S. H. CHEN, JRNL AUTH 2 W. Y. JENG, W. J. CHUANG JRNL TITL SOLUTION STRUCTURE OF THE DNA-BINDING DOMAIN OF JRNL TITL 2 INTERLEUKIN ENHANCER BINDING FACTOR 1 (FOXK 1 A) JRNL REF PROTEINS: V. 49 543 2002 JRNL REF 2 STRUCT. , FUNCT. , GENET.
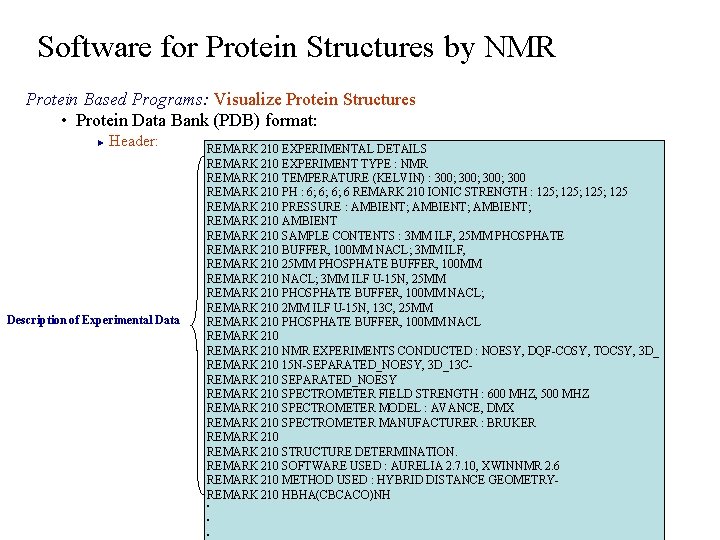
Software for Protein Structures by NMR Protein Based Programs: Visualize Protein Structures • Protein Data Bank (PDB) format: Header: ► Description of Experimental Data REMARK 210 EXPERIMENTAL DETAILS REMARK 210 EXPERIMENT TYPE : NMR REMARK 210 TEMPERATURE (KELVIN) : 300; 300 REMARK 210 PH : 6; 6; 6; 6 REMARK 210 IONIC STRENGTH : 125; 125 REMARK 210 PRESSURE : AMBIENT; REMARK 210 AMBIENT REMARK 210 SAMPLE CONTENTS : 3 MM ILF, 25 MM PHOSPHATE REMARK 210 BUFFER, 100 MM NACL; 3 MM ILF, REMARK 210 25 MM PHOSPHATE BUFFER, 100 MM REMARK 210 NACL; 3 MM ILF U-15 N, 25 MM REMARK 210 PHOSPHATE BUFFER, 100 MM NACL; REMARK 210 2 MM ILF U-15 N, 13 C, 25 MM REMARK 210 PHOSPHATE BUFFER, 100 MM NACL REMARK 210 NMR EXPERIMENTS CONDUCTED : NOESY, DQF-COSY, TOCSY, 3 D_ REMARK 210 15 N-SEPARATED_NOESY, 3 D_13 C- REMARK 210 SEPARATED_NOESY REMARK 210 SPECTROMETER FIELD STRENGTH : 600 MHZ, 500 MHZ REMARK 210 SPECTROMETER MODEL : AVANCE, DMX REMARK 210 SPECTROMETER MANUFACTURER : BRUKER REMARK 210 STRUCTURE DETERMINATION. REMARK 210 SOFTWARE USED : AURELIA 2. 7. 10, XWINNMR 2. 6 REMARK 210 METHOD USED : HYBRID DISTANCE GEOMETRY- REMARK 210 HBHA(CBCACO)NH . . .
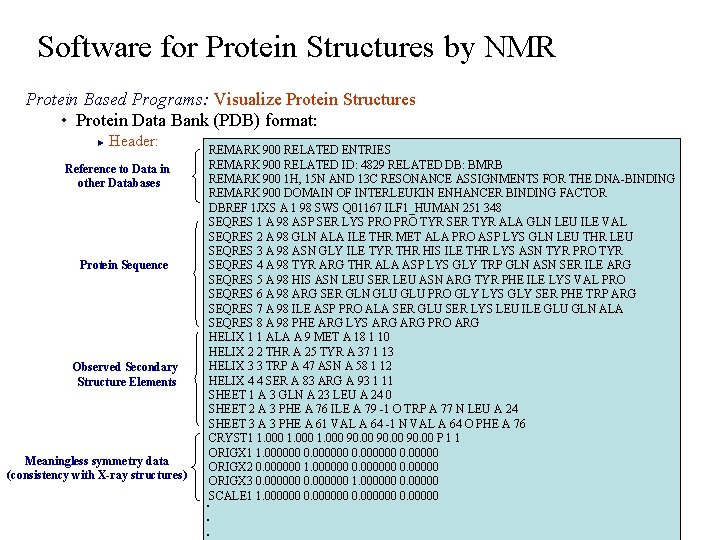
Software for Protein Structures by NMR Protein Based Programs: Visualize Protein Structures • Protein Data Bank (PDB) format: Header: ► Reference to Data in other Databases Protein Sequence Observed Secondary Structure Elements Meaningless symmetry data (consistency with X-ray structures) REMARK 900 RELATED ENTRIES REMARK 900 RELATED ID: 4829 RELATED DB: BMRB REMARK 900 1 H, 15 N AND 13 C RESONANCE ASSIGNMENTS FOR THE DNA-BINDING REMARK 900 DOMAIN OF INTERLEUKIN ENHANCER BINDING FACTOR DBREF 1 JXS A 1 98 SWS Q 01167 ILF 1_HUMAN 251 348 SEQRES 1 A 98 ASP SER LYS PRO TYR SER TYR ALA GLN LEU ILE VAL SEQRES 2 A 98 GLN ALA ILE THR MET ALA PRO ASP LYS GLN LEU THR LEU SEQRES 3 A 98 ASN GLY ILE TYR THR HIS ILE THR LYS ASN TYR PRO TYR SEQRES 4 A 98 TYR ARG THR ALA ASP LYS GLY TRP GLN ASN SER ILE ARG SEQRES 5 A 98 HIS ASN LEU SER LEU ASN ARG TYR PHE ILE LYS VAL PRO SEQRES 6 A 98 ARG SER GLN GLU PRO GLY LYS GLY SER PHE TRP ARG SEQRES 7 A 98 ILE ASP PRO ALA SER GLU SER LYS LEU ILE GLU GLN ALA SEQRES 8 A 98 PHE ARG LYS ARG PRO ARG HELIX 1 1 ALA A 9 MET A 18 1 10 HELIX 2 2 THR A 25 TYR A 37 1 13 HELIX 3 3 TRP A 47 ASN A 58 1 12 HELIX 4 4 SER A 83 ARG A 93 1 11 SHEET 1 A 3 GLN A 23 LEU A 24 0 SHEET 2 A 3 PHE A 76 ILE A 79 -1 O TRP A 77 N LEU A 24 SHEET 3 A 3 PHE A 61 VAL A 64 -1 N VAL A 64 O PHE A 76 CRYST 1 1. 000 90. 00 P 1 1 ORIGX 1 1. 000000 0. 00000 ORIGX 2 0. 000000 1. 000000 0. 00000 ORIGX 3 0. 000000 1. 000000 0. 00000 SCALE 1 1. 000000 0. 00000 . . .
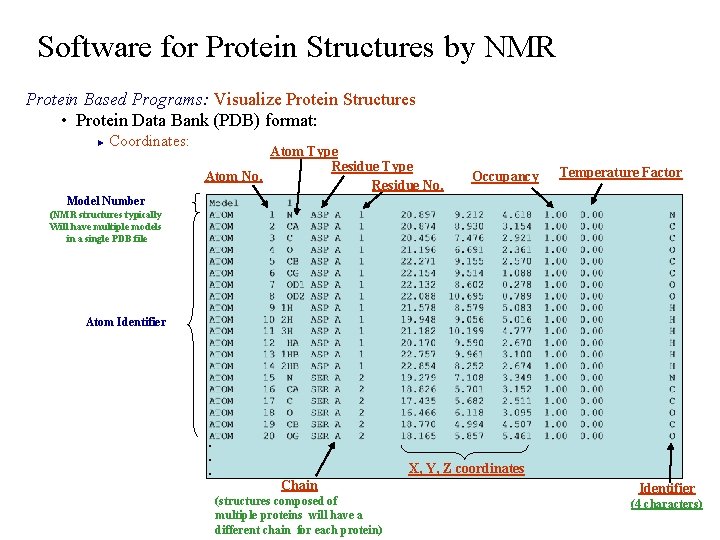
Software for Protein Structures by NMR Protein Based Programs: Visualize Protein Structures • Protein Data Bank (PDB) format: Coordinates: ► Atom Type Residue Type Atom No. Residue No. Occupancy Temperature Factor Model Number (NMR structures typically Will have multiple models in a single PDB file Atom Identifier . . . X, Y, Z coordinates Chain (structures composed of multiple proteins will have a different chain for each protein) Identifier (4 characters)
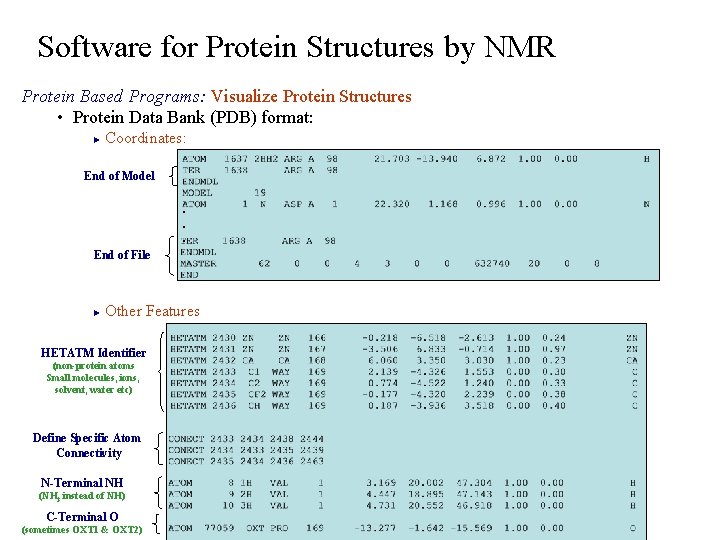
Software for Protein Structures by NMR Protein Based Programs: Visualize Protein Structures • Protein Data Bank (PDB) format: Coordinates: ► End of Model. . . End of File Other Features ► HETATM Identifier (non-protein atoms Small molecules, ions, solvent, water etc) Define Specific Atom Connectivity N-Terminal NH (NH 3 instead of NH) C-Terminal O (sometimes OXT 1 & OXT 2)
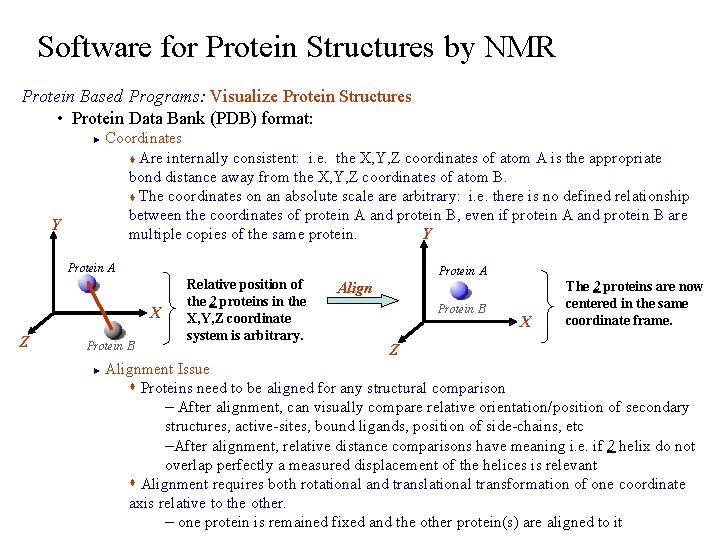
Software for Protein Structures by NMR Protein Based Programs: Visualize Protein Structures • Protein Data Bank (PDB) format: Coordinates t Are internally consistent: i. e. the X, Y, Z coordinates of atom A is the appropriate bond distance away from the X, Y, Z coordinates of atom B. t The coordinates on an absolute scale arbitrary: i. e. there is no defined relationship between the coordinates of protein A and protein B, even if protein A and protein B are Y multiple copies of the same protein. ► Y Protein A X Z Protein B Relative position of the 2 proteins in the X, Y, Z coordinate system is arbitrary. Protein A Align Protein B X The 2 proteins are now centered in the same coordinate frame. Z Alignment Issue s Proteins need to be aligned for any structural comparison – After alignment, can visually compare relative orientation/position of secondary structures, active-sites, bound ligands, position of side-chains, etc –After alignment, relative distance comparisons have meaning i. e. if 2 helix do not overlap perfectly a measured displacement of the helices is relevant s Alignment requires both rotational and translational transformation of one coordinate axis relative to the other. – one protein is remained fixed and the other protein(s) are aligned to it ►
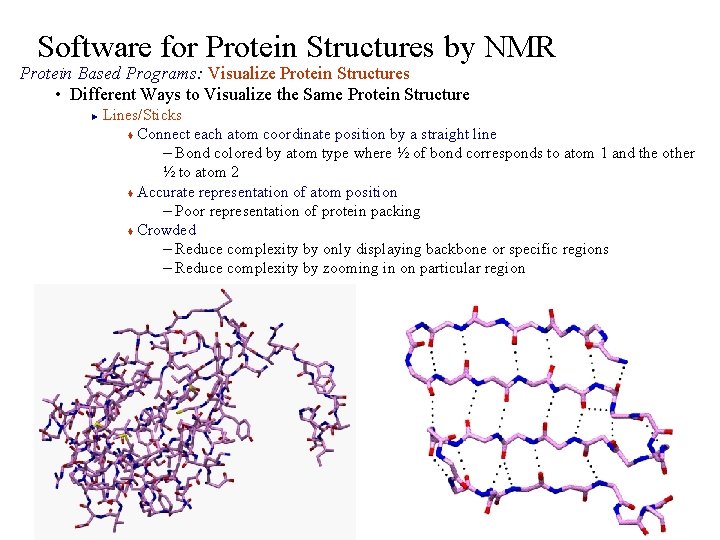
Software for Protein Structures by NMR Protein Based Programs: Visualize Protein Structures • Different Ways to Visualize the Same Protein Structure Lines/Sticks t Connect each atom coordinate position by a straight line – Bond colored by atom type where ½ of bond corresponds to atom 1 and the other ½ to atom 2 t Accurate representation of atom position – Poor representation of protein packing t Crowded – Reduce complexity by only displaying backbone or specific regions – Reduce complexity by zooming in on particular region ►
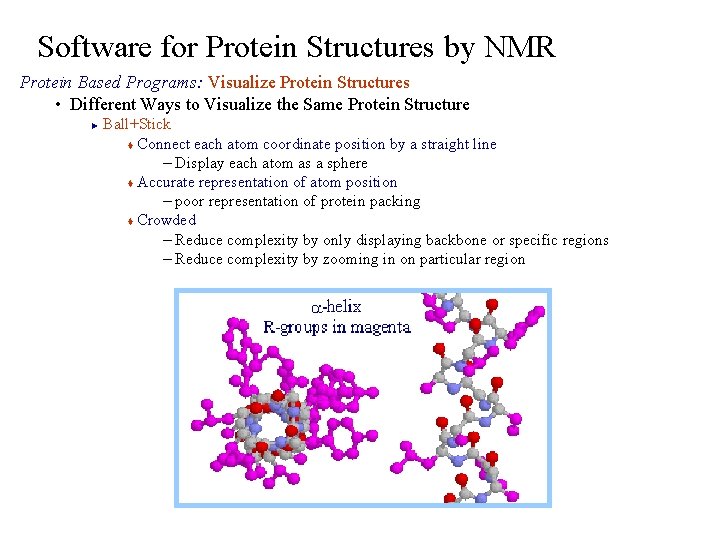
Software for Protein Structures by NMR Protein Based Programs: Visualize Protein Structures • Different Ways to Visualize the Same Protein Structure Ball+Stick t Connect each atom coordinate position by a straight line – Display each atom as a sphere t Accurate representation of atom position – poor representation of protein packing t Crowded – Reduce complexity by only displaying backbone or specific regions – Reduce complexity by zooming in on particular region ►
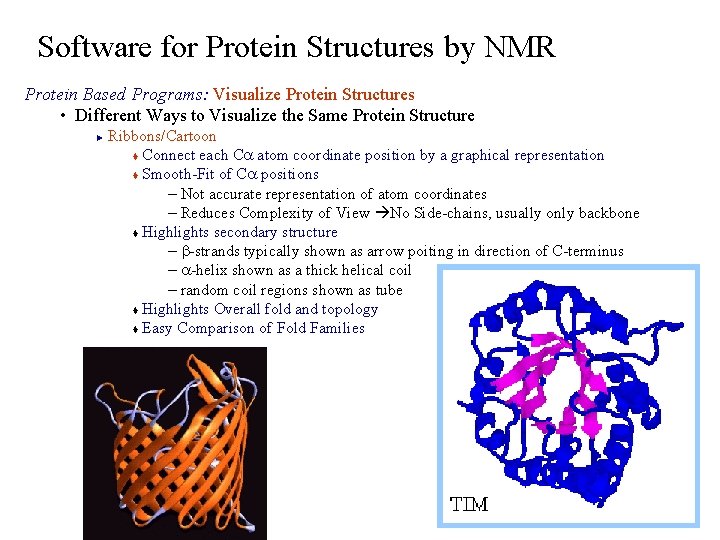
Software for Protein Structures by NMR Protein Based Programs: Visualize Protein Structures • Different Ways to Visualize the Same Protein Structure Ribbons/Cartoon t Connect each Ca atom coordinate position by a graphical representation t Smooth-Fit of Ca positions – Not accurate representation of atom coordinates – Reduces Complexity of View No Side-chains, usually only backbone t Highlights secondary structure – b-strands typically shown as arrow poiting in direction of C-terminus – a-helix shown as a thick helical coil – random coil regions shown as tube t Highlights Overall fold and topology t Easy Comparison of Fold Families ►
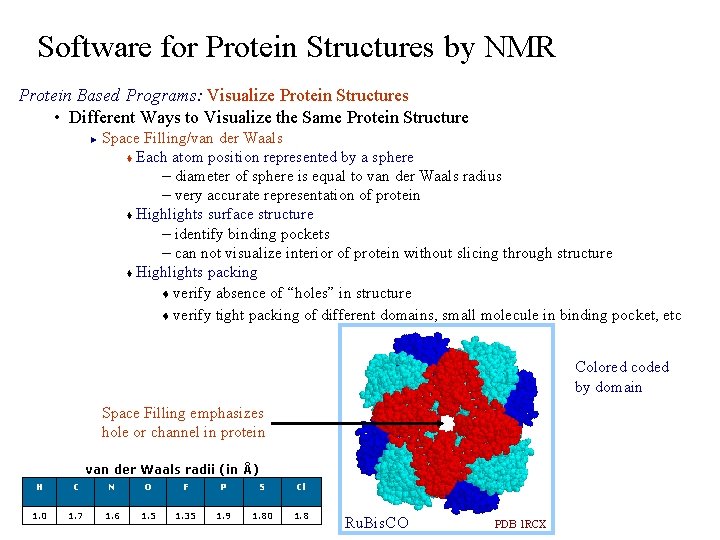
Software for Protein Structures by NMR Protein Based Programs: Visualize Protein Structures • Different Ways to Visualize the Same Protein Structure Space Filling/van der Waals t Each atom position represented by a sphere – diameter of sphere is equal to van der Waals radius – very accurate representation of protein t Highlights surface structure – identify binding pockets – can not visualize interior of protein without slicing through structure t Highlights packing t verify absence of “holes” in structure t verify tight packing of different domains, small molecule in binding pocket, etc ► Colored coded by domain Space Filling emphasizes hole or channel in protein van der Waals radii (in Å) H C N O F P S Cl 1. 0 1. 7 1. 6 1. 5 1. 35 1. 9 1. 80 1. 8
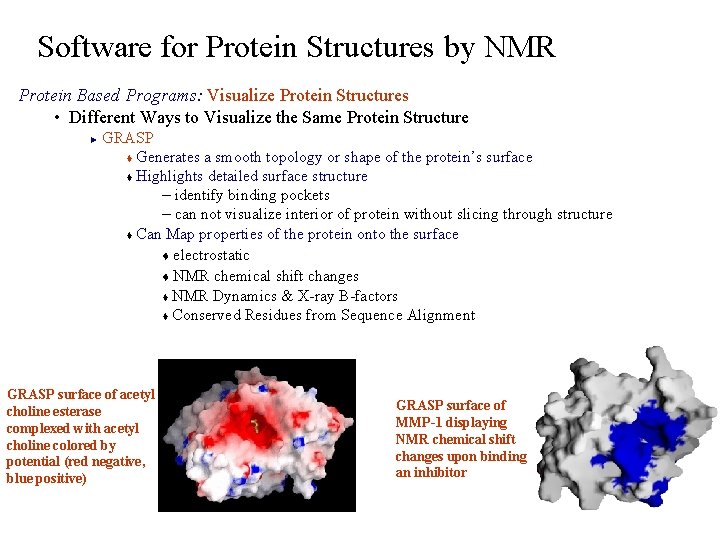
Software for Protein Structures by NMR Protein Based Programs: Visualize Protein Structures • Different Ways to Visualize the Same Protein Structure GRASP t Generates a smooth topology or shape of the protein’s surface t Highlights detailed surface structure – identify binding pockets – can not visualize interior of protein without slicing through structure t Can Map properties of the protein onto the surface t electrostatic t NMR chemical shift changes t NMR Dynamics & X-ray B-factors t Conserved Residues from Sequence Alignment ► GRASP surface of acetyl choline esterase complexed with acetyl choline colored by potential (red negative, blue positive) GRASP surface of MMP-1 displaying NMR chemical shift changes upon binding an inhibitor
Software for Protein Structures by NMR Protein Based Programs: Evaluate Protein Structures • Compare a new protein structure against standard parameters or values standard values or trends are ascertained from analysis of high quality, high resolution structures in the PDB ► typical features as we discussed in the introduction to protein structures ► PROCHECK t A common program used by PDB to validate deposited structures t Assesses the "stereochemical quality" of a given protein structure – reads a PDB formated file – generates 10 output postscript files – analyzes f, y, c 1, c 2 torsion angles, bond lengths bond angles – analyzes “bad contacts” atoms too close by van der waals radius – analyzes hydrogen bond energy – analyzes G-factor ► § Compares bond lengths and bond angles to database of standard small molecule values – Provides overall and per residue analyses – Identifies distorted geometry
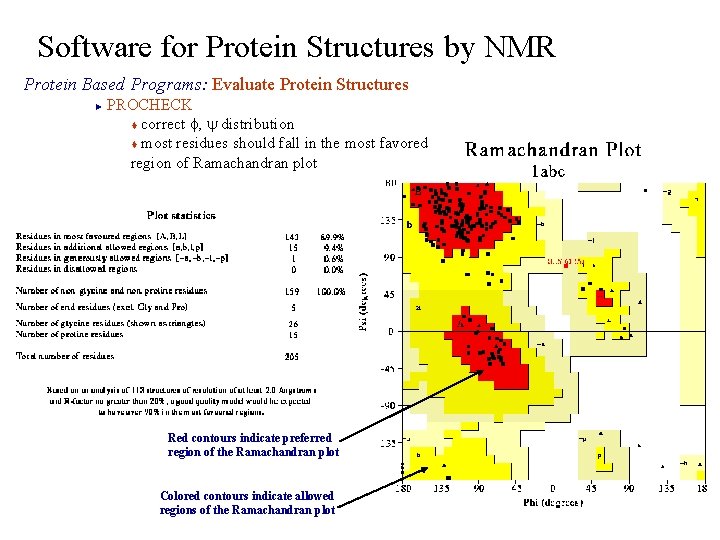
Software for Protein Structures by NMR Protein Based Programs: Evaluate Protein Structures PROCHECK t correct f, y distribution t most residues should fall in the most favored region of Ramachandran plot ► Red contours indicate preferred region of the Ramachandran plot Colored contours indicate allowed regions of the Ramachandran plot
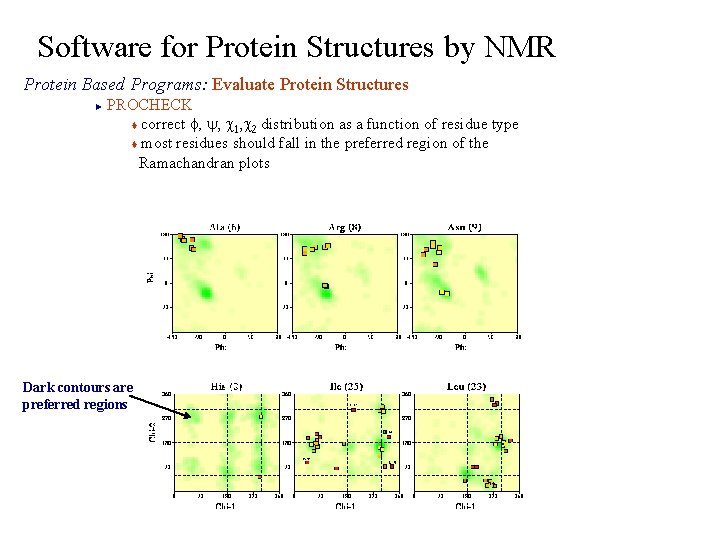
Software for Protein Structures by NMR Protein Based Programs: Evaluate Protein Structures PROCHECK t correct f, y, c 1, c 2 distribution as a function of residue type t most residues should fall in the preferred region of the Ramachandran plots ► Dark contours are preferred regions
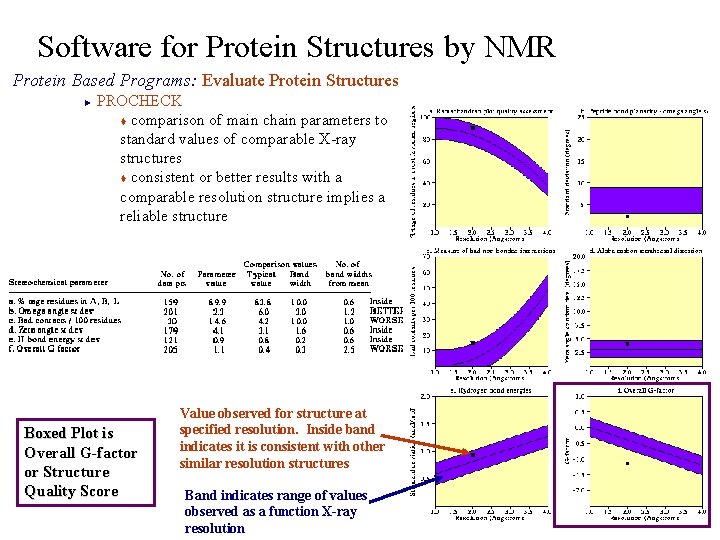
Software for Protein Structures by NMR Protein Based Programs: Evaluate Protein Structures PROCHECK t comparison of main chain parameters to standard values of comparable X-ray structures t consistent or better results with a comparable resolution structure implies a reliable structure ► Boxed Plot is Overall G-factor or Structure Quality Score Value observed for structure at specified resolution. Inside band indicates it is consistent with other similar resolution structures Band indicates range of values observed as a function X-ray resolution
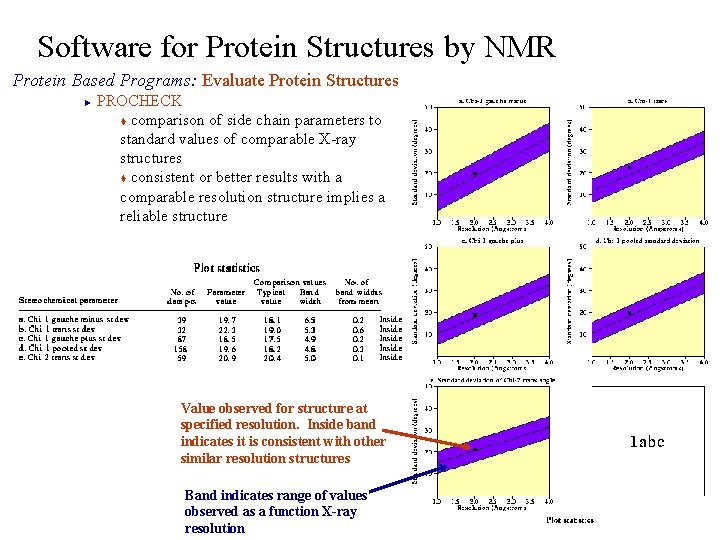
Software for Protein Structures by NMR Protein Based Programs: Evaluate Protein Structures PROCHECK t comparison of side chain parameters to standard values of comparable X-ray structures t consistent or better results with a comparable resolution structure implies a reliable structure ► Value observed for structure at specified resolution. Inside band indicates it is consistent with other similar resolution structures Band indicates range of values observed as a function X-ray resolution
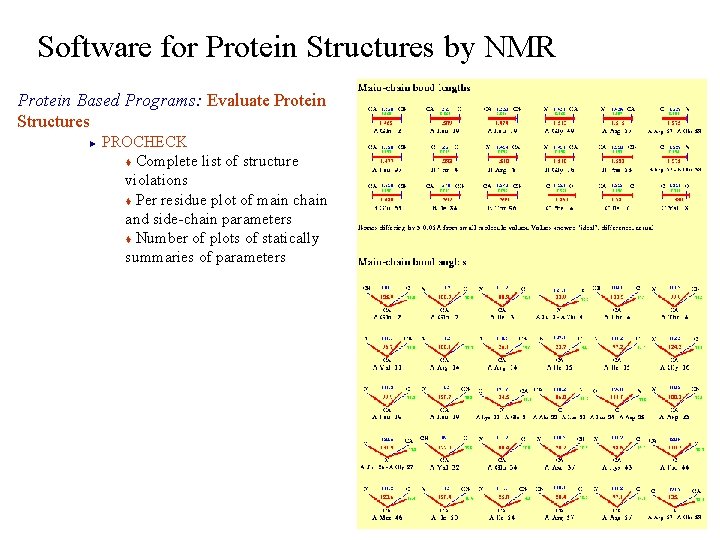
Software for Protein Structures by NMR Protein Based Programs: Evaluate Protein Structures PROCHECK t Complete list of structure violations t Per residue plot of main chain and side-chain parameters t Number of plots of statically summaries of parameters ►
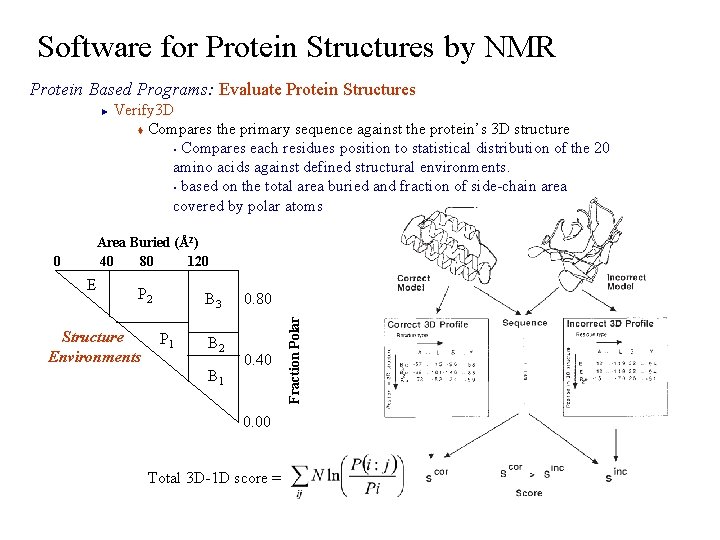
Software for Protein Structures by NMR Protein Based Programs: Evaluate Protein Structures Verify 3 D t Compares the primary sequence against the protein’s 3 D structure § Compares each residues position to statistical distribution of the 20 amino acids against defined structural environments. § based on the total area buried and fraction of side-chain area covered by polar atoms ► E P 2 Structure Environments B 3 P 1 B 2 B 1 0. 80 0. 40 0. 00 Total 3 D-1 D score = Fraction Polar 0 Area Buried (Å2) 120 40 80
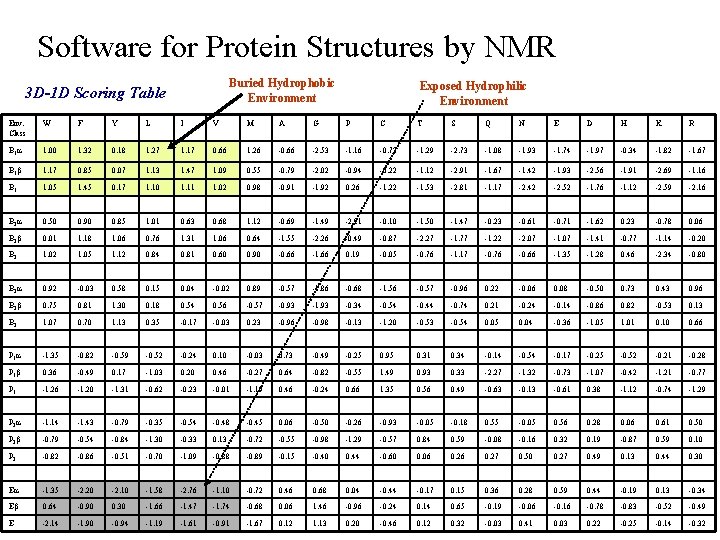
Software for Protein Structures by NMR Buried Hydrophobic Environment 3 D-1 D Scoring Table Exposed Hydrophilic Environment Env. Class W F Y L I V M A G P C T S Q N E D H K R B 1 a 1. 00 1. 32 0. 18 1. 27 1. 17 0. 66 1. 26 -0. 66 -2. 53 -1. 16 -0. 73 -1. 29 -2. 73 -1. 08 -1. 93 -1. 74 -1. 97 -0. 34 -1. 82 -1. 67 B 1 b 1. 17 0. 85 0. 07 1. 13 1. 47 1. 09 0. 55 -0. 79 -2. 02 -0. 94 -0. 22 -1. 12 -2. 91 -1. 67 -1. 42 -1. 93 -2. 56 -1. 91 -2. 69 -1. 16 B 1 1. 05 1. 45 0. 17 1. 10 1. 11 1. 02 0. 98 -0. 91 -1. 92 0. 26 -1. 22 -1. 53 -2. 81 -1. 17 -2. 42 -2. 52 -1. 76 -1. 12 -2. 59 -2. 16 B 2 a 0. 50 0. 90 0. 85 1. 01 0. 63 0. 68 1. 12 -0. 69 -1. 49 -2. 21 -0. 10 -1. 50 -1. 47 -0. 23 -0. 61 -0. 71 -1. 62 0. 23 -0. 78 0. 06 B 2 b 0. 01 1. 18 1. 06 0. 76 1. 31 1. 06 0. 64 -1. 55 -2. 26 -0. 49 -0. 87 -2. 27 -1. 77 -1. 22 -2. 07 -1. 41 -0. 77 -1. 14 -0. 20 B 2 1. 05 1. 12 0. 84 0. 81 0. 60 0. 90 -0. 66 -1. 66 0. 19 -0. 05 -0. 76 -1. 17 -0. 76 -0. 66 -1. 35 -1. 28 0. 46 -2. 34 -0. 80 B 3 a 0. 92 -0. 03 0. 58 0. 15 0. 04 -0. 02 0. 89 -0. 57 -1. 86 -0. 68 -1. 56 -0. 57 -0. 96 0. 22 -0. 06 0. 08 -0. 50 0. 73 0. 43 0. 96 B 3 b 0. 75 0. 81 1. 30 0. 18 0. 54 0. 56 -0. 57 -0. 93 -1. 93 -0. 34 -0. 54 -0. 44 -0. 74 0. 21 -0. 24 -0. 14 -0. 86 0. 82 -0. 53 0. 13 B 3 1. 07 0. 70 1. 13 0. 35 -0. 17 -0. 03 0. 23 -0. 96 -0. 98 -0. 13 -1. 20 -0. 53 -0. 54 0. 05 0. 04 -0. 36 -1. 05 1. 01 0. 10 0. 66 P 1 a -1. 35 -0. 82 -0. 59 -0. 52 -0. 24 0. 10 -0. 03 0. 73 -0. 49 -0. 25 0. 95 0. 31 0. 34 -0. 14 -0. 54 -0. 17 -0. 25 -0. 52 -0. 21 -0. 28 P 1 b 0. 36 -0. 49 0. 17 -1. 03 0. 20 0. 46 -0. 27 0. 64 -0. 82 -0. 55 1. 49 0. 93 0. 33 -2. 27 -1. 32 -0. 73 -1. 07 -0. 42 -1. 21 -0. 77 P 1 -1. 26 -1. 20 -1. 31 -0. 62 -0. 23 -0. 01 -1. 19 0. 46 -0. 24 0. 66 1. 35 0. 56 0. 49 -0. 63 -0. 13 -0. 61 0. 38 -1. 12 -0. 74 -1. 29 P 2 a -1. 14 -1. 43 -0. 79 -0. 35 -0. 54 -0. 48 -0. 45 0. 06 -0. 50 -0. 26 -0. 93 -0. 05 -0. 18 0. 55 -0. 05 0. 56 0. 28 0. 06 0. 61 0. 50 P 2 b -0. 79 -0. 54 -0. 84 -1. 30 -0. 33 0. 13 -0. 72 -0. 55 -0. 98 -1. 29 -0. 57 0. 84 0. 59 -0. 08 -0. 16 0. 32 0. 19 -0. 87 0. 59 0. 10 P 2 -0. 86 -0. 51 -0. 70 -1. 09 -0. 88 -0. 89 -0. 15 -0. 40 0. 44 -0. 60 0. 06 0. 27 0. 50 0. 27 0. 49 0. 13 0. 44 0. 30 Ea -1. 35 -2. 20 -2. 10 -1. 58 -2. 76 -1. 10 -0. 72 0. 46 0. 68 0. 04 -0. 44 -0. 17 0. 15 0. 36 0. 28 0. 59 0. 44 -0. 19 0. 13 -0. 34 Eb 0. 64 -0. 90 0. 30 -1. 66 -1. 47 -1. 74 -0. 68 0. 06 1. 46 -0. 96 -0. 24 0. 14 0. 65 -0. 19 -0. 06 -0. 16 -0. 78 -0. 83 -0. 52 -0. 49 E -2. 14 -1. 90 -0. 94 -1. 19 -1. 61 -0. 91 -1. 67 0. 12 1. 13 0. 20 -0. 46 0. 12 0. 32 -0. 03 0. 41 0. 03 0. 22 -0. 25 -0. 14 -0. 32
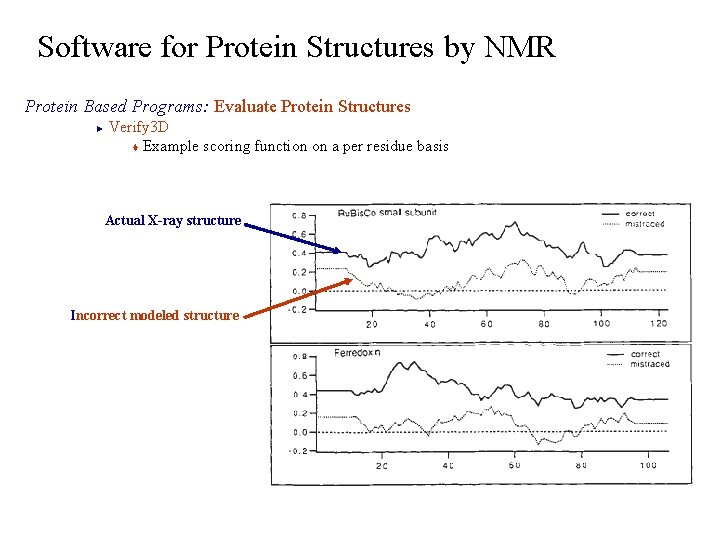
Software for Protein Structures by NMR Protein Based Programs: Evaluate Protein Structures Verify 3 D t Example scoring function on a per residue basis ► Actual X-ray structure Incorrect modeled structure
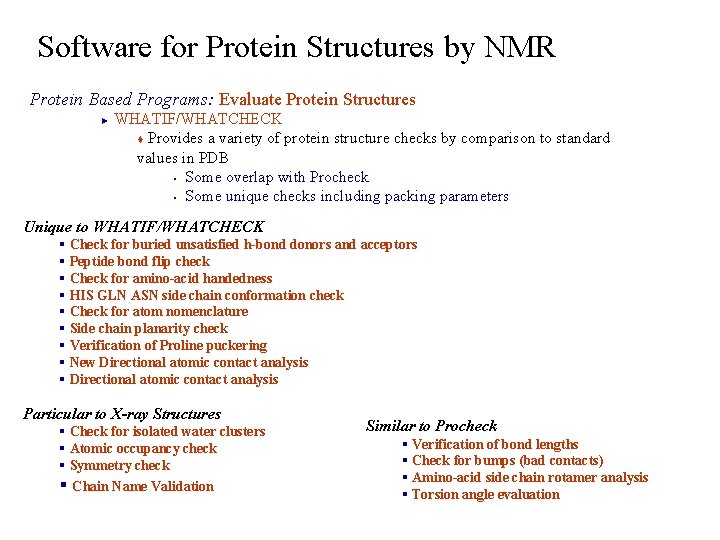
Software for Protein Structures by NMR Protein Based Programs: Evaluate Protein Structures WHATIF/WHATCHECK t Provides a variety of protein structure checks by comparison to standard values in PDB § Some overlap with Procheck § Some unique checks including packing parameters ► Unique to WHATIF/WHATCHECK § Check for buried unsatisfied h-bond donors and acceptors § Peptide bond flip check § Check for amino-acid handedness § HIS GLN ASN side chain conformation check § Check for atom nomenclature § Side chain planarity check § Verification of Proline puckering § New Directional atomic contact analysis § Directional atomic contact analysis Particular to X-ray Structures § Check for isolated water clusters § Atomic occupancy check § Symmetry check § Chain Name Validation Similar to Procheck § Verification of bond lengths § Check for bumps (bad contacts) § Amino-acid side chain rotamer analysis § Torsion angle evaluation
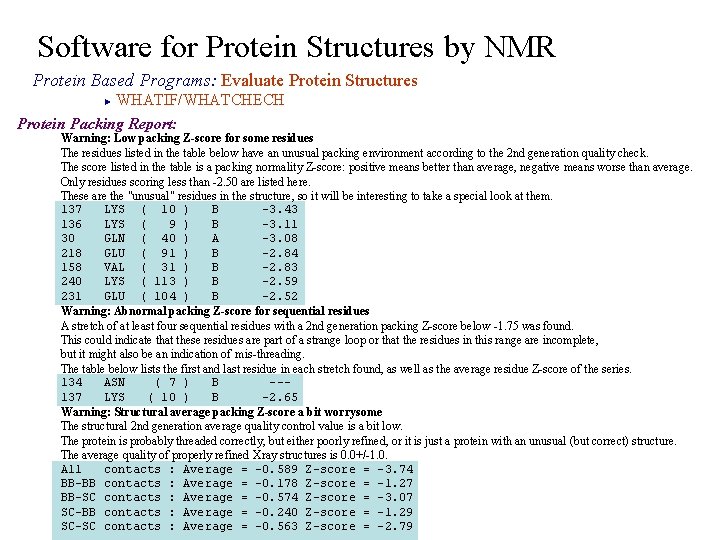
Software for Protein Structures by NMR Protein Based Programs: Evaluate Protein Structures WHATIF/WHATCHECH Protein Packing Report: ► Warning: Low packing Z-score for some residues The residues listed in the table below have an unusual packing environment according to the 2 nd generation quality check. The score listed in the table is a packing normality Z-score: positive means better than average, negative means worse than average. Only residues scoring less than -2. 50 are listed here. These are the "unusual" residues in the structure, so it will be interesting to take a special look at them. 137 LYS ( 10 ) B -3. 43 136 LYS ( 9 ) B -3. 11 30 GLN ( 40 ) A -3. 08 218 GLU ( 91 ) B -2. 84 158 VAL ( 31 ) B -2. 83 240 LYS ( 113 ) B -2. 59 231 GLU ( 104 ) B -2. 52 Warning: Abnormal packing Z-score for sequential residues A stretch of at least four sequential residues with a 2 nd generation packing Z-score below -1. 75 was found. This could indicate that these residues are part of a strange loop or that the residues in this range are incomplete, but it might also be an indication of mis-threading. The table below lists the first and last residue in each stretch found, as well as the average residue Z-score of the series. 134 ASN ( 7 ) B --137 LYS ( 10 ) B -2. 65 Warning: Structural average packing Z-score a bit worrysome The structural 2 nd generation average quality control value is a bit low. The protein is probably threaded correctly, but either poorly refined, or it is just a protein with an unusual (but correct) structure. The average quality of properly refined Xray structures is 0. 0+/-1. 0. All contacts : Average = -0. 589 Z-score = -3. 74 BB-BB contacts : Average = -0. 178 Z-score = -1. 27 BB-SC contacts : Average = -0. 574 Z-score = -3. 07 SC-BB contacts : Average = -0. 240 Z-score = -1. 29 SC-SC contacts : Average = -0. 563 Z-score = -2. 79
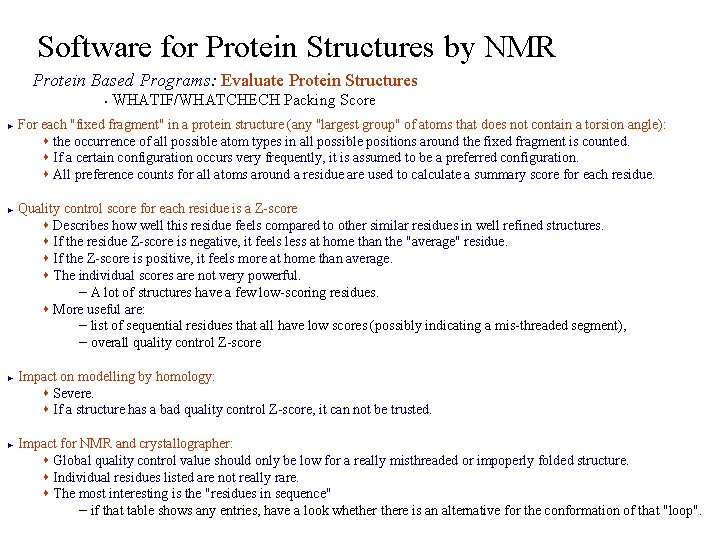
Software for Protein Structures by NMR Protein Based Programs: Evaluate Protein Structures WHATIF/WHATCHECH Packing Score § For each "fixed fragment" in a protein structure (any "largest group" of atoms that does not contain a torsion angle): s the occurrence of all possible atom types in all possible positions around the fixed fragment is counted. s If a certain configuration occurs very frequently, it is assumed to be a preferred configuration. s All preference counts for all atoms around a residue are used to calculate a summary score for each residue. ► Quality control score for each residue is a Z-score s Describes how well this residue feels compared to other similar residues in well refined structures. s If the residue Z-score is negative, it feels less at home than the "average" residue. s If the Z-score is positive, it feels more at home than average. s The individual scores are not very powerful. – A lot of structures have a few low-scoring residues. s More useful are: – list of sequential residues that all have low scores (possibly indicating a mis-threaded segment), – overall quality control Z-score ► ► Impact on modelling by homology: s Severe. s If a structure has a bad quality control Z-score, it can not be trusted. ► Impact for NMR and crystallographer: s Global quality control value should only be low for a really misthreaded or impoperly folded structure. s Individual residues listed are not really rare. s The most interesting is the "residues in sequence" – if that table shows any entries, have a look whethere is an alternative for the conformation of that "loop".
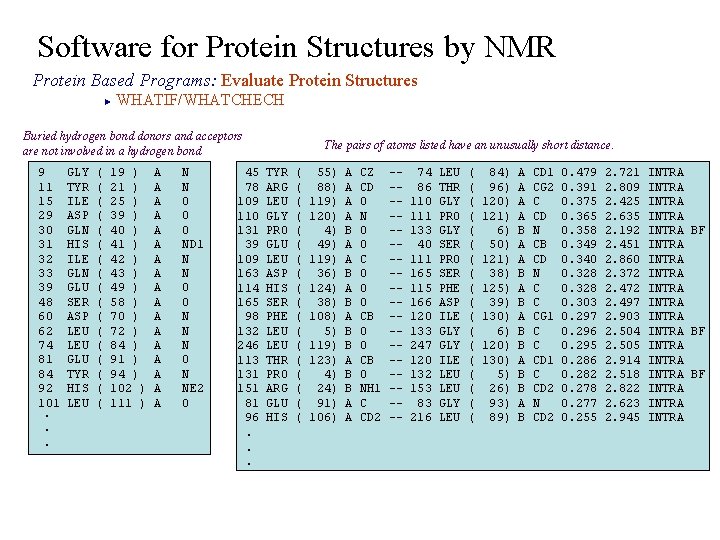
Software for Protein Structures by NMR Protein Based Programs: Evaluate Protein Structures WHATIF/WHATCHECH ► Buried hydrogen bond donors and acceptors are not involved in a hydrogen bond 9 11 15 29 30 31 32 33 39 48 60 62 74 81 84 92 101. . . GLY TYR ILE ASP GLN HIS ILE GLN GLU SER ASP LEU GLU TYR HIS LEU ( ( ( ( ( 19 ) 21 ) 25 ) 39 ) 40 ) 41 ) 42 ) 43 ) 49 ) 58 ) 70 ) 72 ) 84 ) 91 ) 94 ) 102 ) 111 ) A A A A A N N O O O ND 1 N N O O N N N O N NE 2 O 45 78 109 110 131 39 109 163 114 165 98 132 246 113 131 151 81 96. . . The pairs of atoms listed have an unusually short distance. TYR ARG LEU GLY PRO GLU LEU ASP HIS SER PHE LEU THR PRO ARG GLU HIS ( ( ( ( ( 55) 88) 119) 120) 4) 49) 119) 36) 124) 38) 108) 5) 119) 123) 4) 24) 91) 106) A A B A B B A A CZ CD O N O O C O O O CB O NH 1 C CD 2 ---------- 74 86 110 111 133 40 111 165 115 166 120 133 247 120 132 153 83 216 LEU THR GLY PRO GLY SER PRO SER PHE ASP ILE GLY ILE LEU GLY LEU ( ( ( ( ( 84) 96) 120) 121) 6) 50) 121) 38) 125) 39) 130) 6) 120) 130) 5) 26) 93) 89) A A B A B B A B CD 1 CG 2 C CD N CB CD N C C CG 1 C C CD 1 C CD 2 N CD 2 0. 479 0. 391 0. 375 0. 365 0. 358 0. 349 0. 340 0. 328 0. 303 0. 297 0. 296 0. 295 0. 286 0. 282 0. 278 0. 277 0. 255 2. 721 2. 809 2. 425 2. 635 2. 192 2. 451 2. 860 2. 372 2. 497 2. 903 2. 504 2. 505 2. 914 2. 518 2. 822 2. 623 2. 945 INTRA INTRA INTRA BF INTRA INTRA
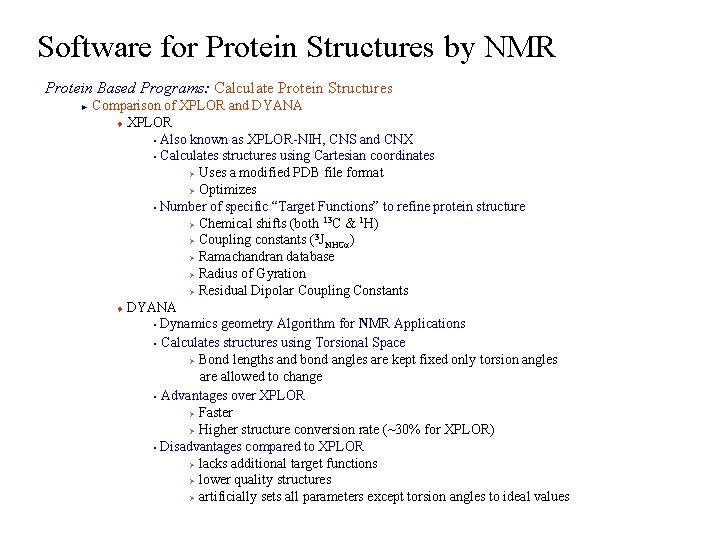
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures Comparison of XPLOR and DYANA t XPLOR § Also known as XPLOR-NIH, CNS and CNX § Calculates structures using Cartesian coordinates Ø Uses a modified PDB file format Ø Optimizes § Number of specific “Target Functions” to refine protein structure 13 1 Ø Chemical shifts (both C & H) 3 Ø Coupling constants ( JNHCa) Ø Ramachandran database Ø Radius of Gyration Ø Residual Dipolar Coupling Constants t DYANA § Dynamics geometry Algorithm for NMR Applications § Calculates structures using Torsional Space Ø Bond lengths and bond angles are kept fixed only torsion angles are allowed to change § Advantages over XPLOR Ø Faster Ø Higher structure conversion rate (~30% for XPLOR) § Disadvantages compared to XPLOR Ø lacks additional target functions Ø lower quality structures Ø artificially sets all parameters except torsion angles to ideal values ►
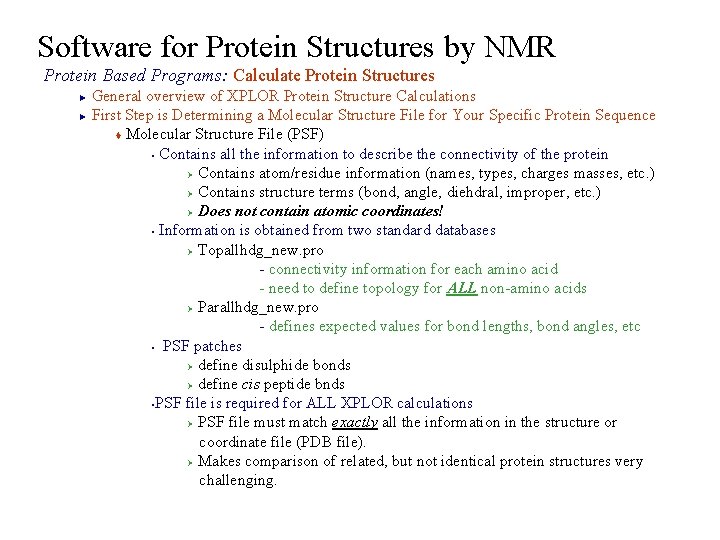
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations ► First Step is Determining a Molecular Structure File for Your Specific Protein Sequence t Molecular Structure File (PSF) § Contains all the information to describe the connectivity of the protein Ø Contains atom/residue information (names, types, charges masses, etc. ) Ø Contains structure terms (bond, angle, diehdral, improper, etc. ) Ø Does not contain atomic coordinates! § Information is obtained from two standard databases Ø Topallhdg_new. pro - connectivity information for each amino acid - need to define topology for ALL non-amino acids Ø Parallhdg_new. pro - defines expected values for bond lengths, bond angles, etc § PSF patches Ø define disulphide bonds Ø define cis peptide bnds §PSF file is required for ALL XPLOR calculations Ø PSF file must match exactly all the information in the structure or coordinate file (PDB file). Ø Makes comparison of related, but not identical protein structures very challenging. ►
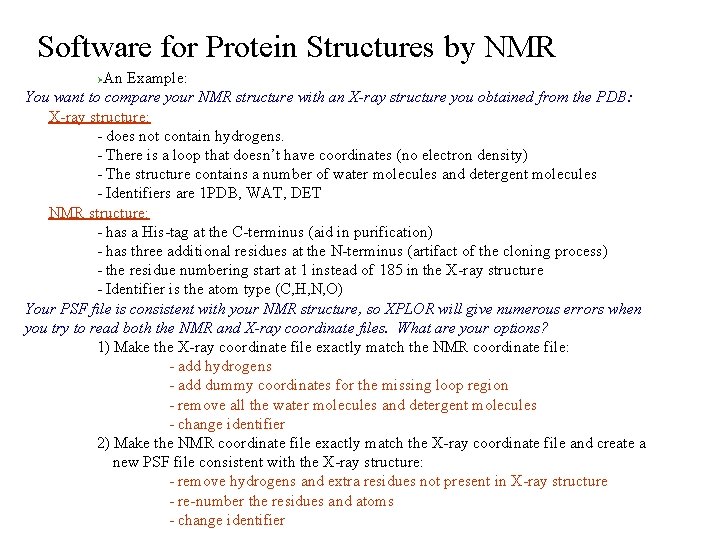
Software for Protein Structures by NMR An Example: You want to compare your NMR structure with an X-ray structure you obtained from the PDB: X-ray structure: - does not contain hydrogens. - There is a loop that doesn’t have coordinates (no electron density) - The structure contains a number of water molecules and detergent molecules - Identifiers are 1 PDB, WAT, DET NMR structure: - has a His-tag at the C-terminus (aid in purification) - has three additional residues at the N-terminus (artifact of the cloning process) - the residue numbering start at 1 instead of 185 in the X-ray structure - Identifier is the atom type (C, H, N, O) Your PSF file is consistent with your NMR structure, so XPLOR will give numerous errors when you try to read both the NMR and X-ray coordinate files. What are your options? 1) Make the X-ray coordinate file exactly match the NMR coordinate file: - add hydrogens - add dummy coordinates for the missing loop region - remove all the water molecules and detergent molecules - change identifier 2) Make the NMR coordinate file exactly match the X-ray coordinate file and create a new PSF file consistent with the X-ray structure: - remove hydrogens and extra residues not present in X-ray structure - re-number the residues and atoms - change identifier Ø
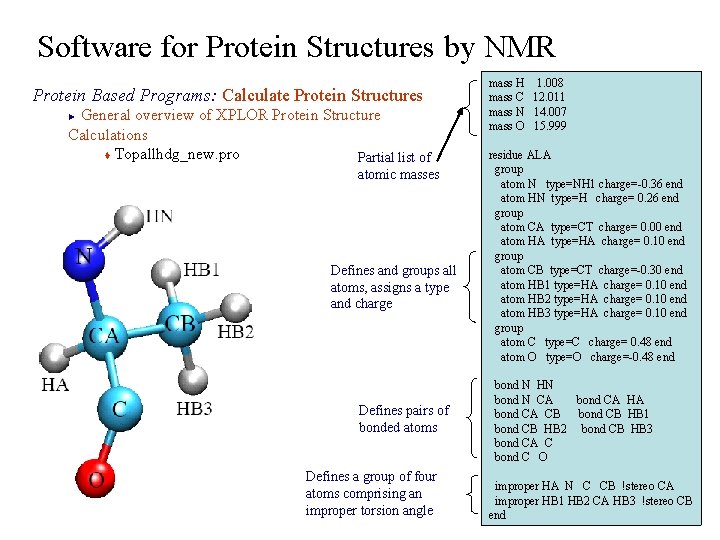
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations t Topallhdg_new. pro Partial list of ► atomic masses Defines and groups all atoms, assigns a type and charge Defines pairs of bonded atoms Defines a group of four atoms comprising an improper torsion angle mass H 1. 008 mass C 12. 011 mass N 14. 007 mass O 15. 999 residue ALA group atom N type=NH 1 charge=-0. 36 end atom HN type=H charge= 0. 26 end group atom CA type=CT charge= 0. 00 end atom HA type=HA charge= 0. 10 end group atom CB type=CT charge=-0. 30 end atom HB 1 type=HA charge= 0. 10 end atom HB 2 type=HA charge= 0. 10 end atom HB 3 type=HA charge= 0. 10 end group atom C type=C charge= 0. 48 end atom O type=O charge=-0. 48 end bond N HN bond N CA bond CA HA bond CA CB bond CB HB 1 bond CB HB 2 bond CB HB 3 bond CA C bond C O improper HA N C CB !stereo CA improper HB 1 HB 2 CA HB 3 !stereo CB end
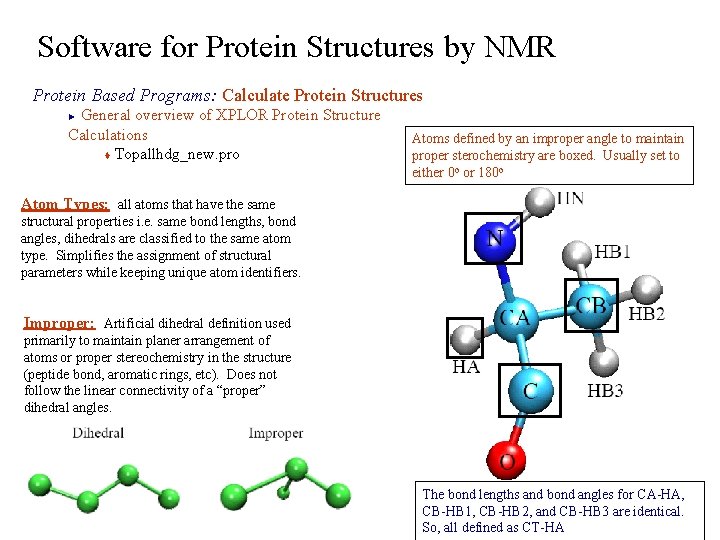
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations t Topallhdg_new. pro ► Atoms defined by an improper angle to maintain proper sterochemistry are boxed. Usually set to either 0 o or 180 o Atom Types: all atoms that have the same structural properties i. e. same bond lengths, bond angles, dihedrals are classified to the same atom type. Simplifies the assignment of structural parameters while keeping unique atom identifiers. Improper: Artificial dihedral definition used primarily to maintain planer arrangement of atoms or proper stereochemistry in the structure (peptide bond, aromatic rings, etc). Does not follow the linear connectivity of a “proper” dihedral angles. The bond lengths and bond angles for CA-HA, CB-HB 1, CB-HB 2, and CB-HB 3 are identical. So, all defined as CT-HA
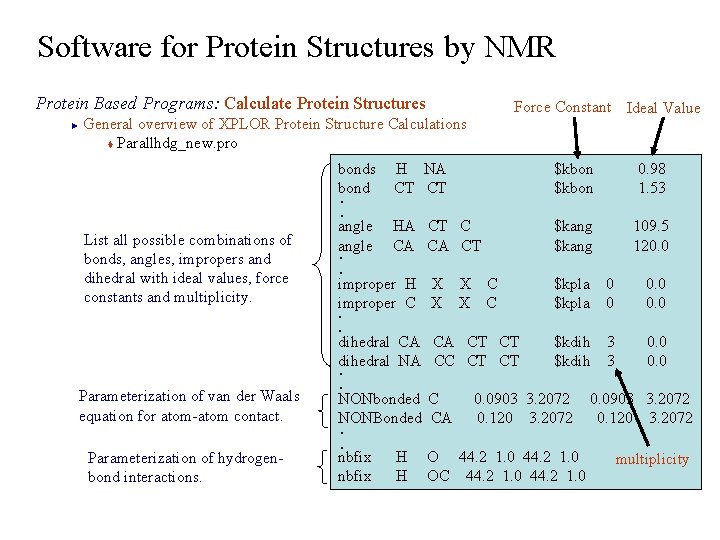
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations t Parallhdg_new. pro Force Constant Ideal Value ► bonds H NA bond CT $kbon 0. 98 $kbon 1. 53 angle HA CT C angle CA CT $kang 109. 5 $kang 120. 0 improper H X X C improper C X X C $kpla 0 0. 0 . . List all possible combinations of bonds, angles, impropers and dihedral with ideal values, force constants and multiplicity. . . dihedral CA CT $kdih 3 0. 0 dihedral NA CC CT $kdih 3 0. 0 Parameterization of van der Waals equation for atom-atom contact. Parameterization of hydrogenbond interactions. . . NONbonded C 0. 0903 3. 2072 0. 0903 3. 2072 NONBonded CA 0. 120 3. 2072. . nbfix H O 44. 2 1. 0 nbfix H OC 44. 2 1. 0 multiplicity
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations t Parallhdg_new. pro t Defining atomic parameters is a very active area of molecular modelling research t. The values in the parameter database come from multiple sources: § X-ray database of high-resolution small molecules § ab initio calculations § experimental observations, IR, Raman, water-ion neutron and X-ray diffraction data, free energy of solvation data, etc ►
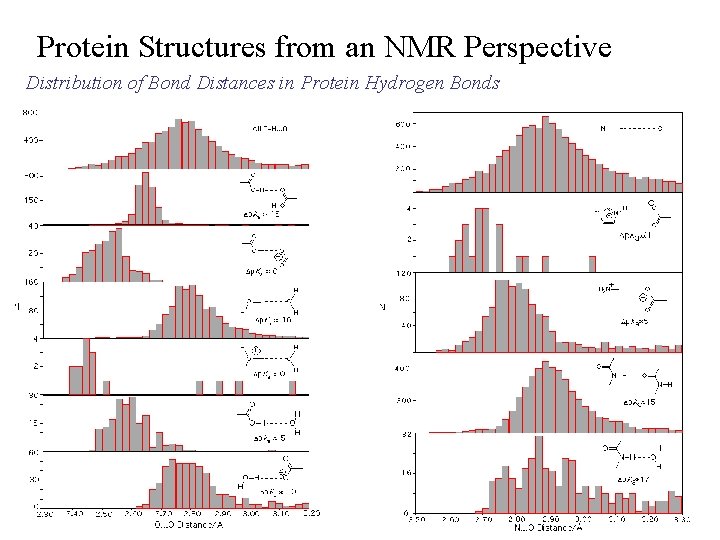
Protein Structures from an NMR Perspective Distribution of Bond Distances in Protein Hydrogen Bonds
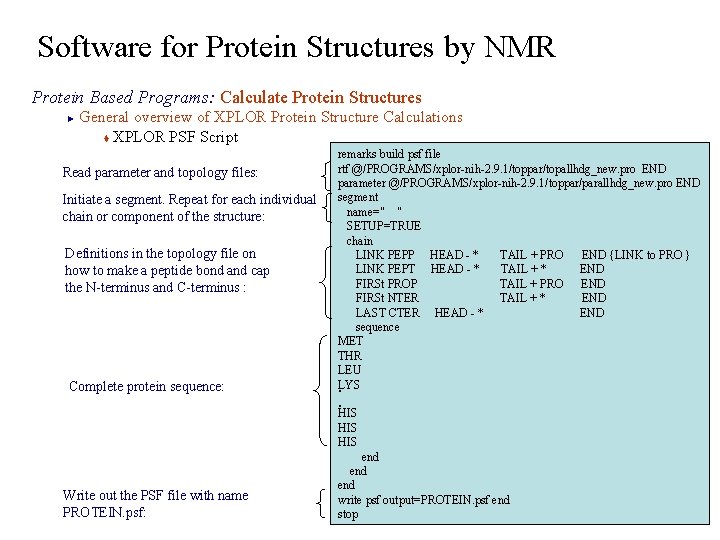
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations t XPLOR PSF Script ► Read parameter and topology files: Initiate a segment. Repeat for each individual chain or component of the structure: Definitions in the topology file on how to make a peptide bond and cap the N-terminus and C-terminus : Complete protein sequence: Write out the PSF file with name PROTEIN. psf: remarks build psf file rtf @/PROGRAMS/xplor-nih-2. 9. 1/toppar/topallhdg_new. pro END parameter @/PROGRAMS/xplor-nih-2. 9. 1/toppar/parallhdg_new. pro END segment name=" " SETUP=TRUE chain LINK PEPP HEAD - * TAIL + PRO END {LINK to PRO } LINK PEPT HEAD - * TAIL + * END FIRSt PROP TAIL + PRO END FIRSt NTER TAIL + * END LAST CTER HEAD - * END sequence MET THR LEU LYS. . HIS HIS end end write psf output=PROTEIN. psf end stop
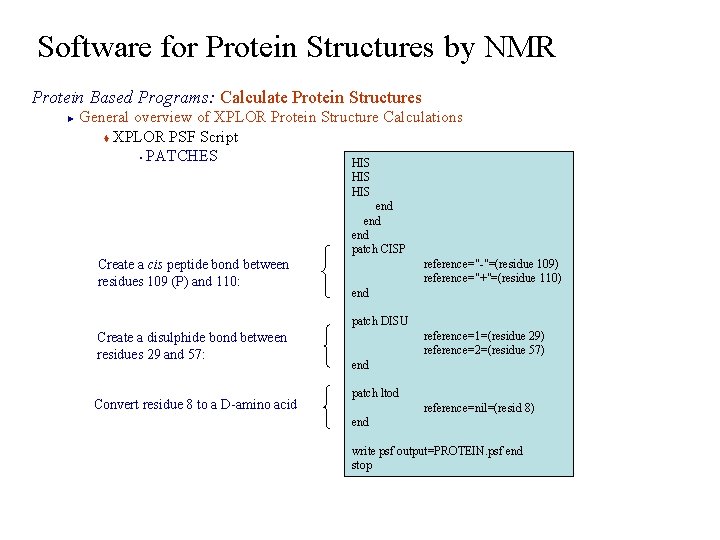
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations t XPLOR PSF Script § PATCHES HIS ► HIS end end patch CISP Create a cis peptide bond between residues 109 (P) and 110: end reference="-"=(residue 109) reference="+"=(residue 110) patch DISU Create a disulphide bond between residues 29 and 57: Convert residue 8 to a D-amino acid reference=1=(residue 29) reference=2=(residue 57) end patch ltod reference=nil=(resid 8) end write psf output=PROTEIN. psf end stop
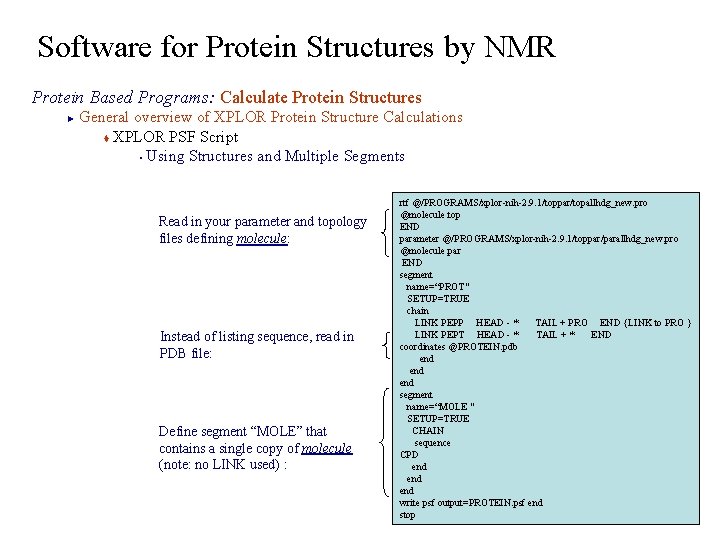
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations t XPLOR PSF Script § Using Structures and Multiple Segments ► Read in your parameter and topology files defining molecule: Instead of listing sequence, read in PDB file: Define segment “MOLE” that contains a single copy of molecule (note: no LINK used) : rtf @/PROGRAMS/xplor-nih-2. 9. 1/toppar/topallhdg_new. pro @molecule. top END parameter @/PROGRAMS/xplor-nih-2. 9. 1/toppar/parallhdg_new. pro @molecule. par END segment name=“PROT" SETUP=TRUE chain LINK PEPP HEAD - * TAIL + PRO END {LINK to PRO } LINK PEPT HEAD - * TAIL + * END coordinates @PROTEIN. pdb end end segment name=“MOLE " SETUP=TRUE CHAIN sequence CPD end end write psf output=PROTEIN. psf end stop
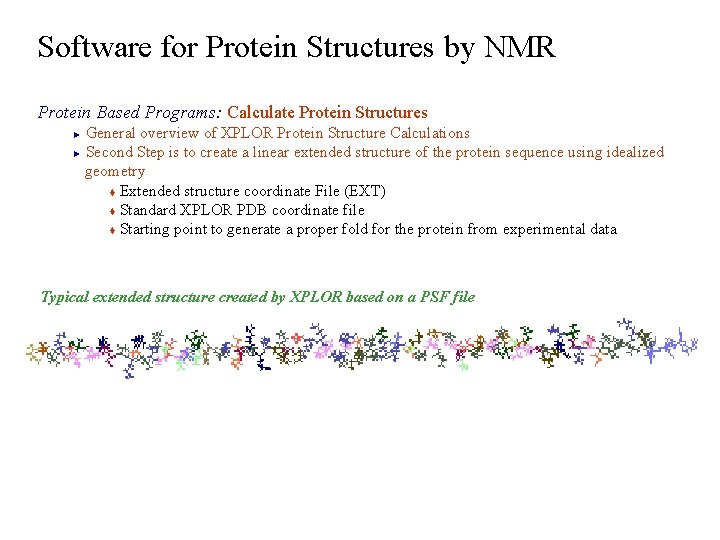
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations ► Second Step is to create a linear extended structure of the protein sequence using idealized geometry t Extended structure coordinate File (EXT) t Standard XPLOR PDB coordinate file t Starting point to generate a proper fold for the protein from experimental data ► Typical extended structure created by XPLOR based on a PSF file
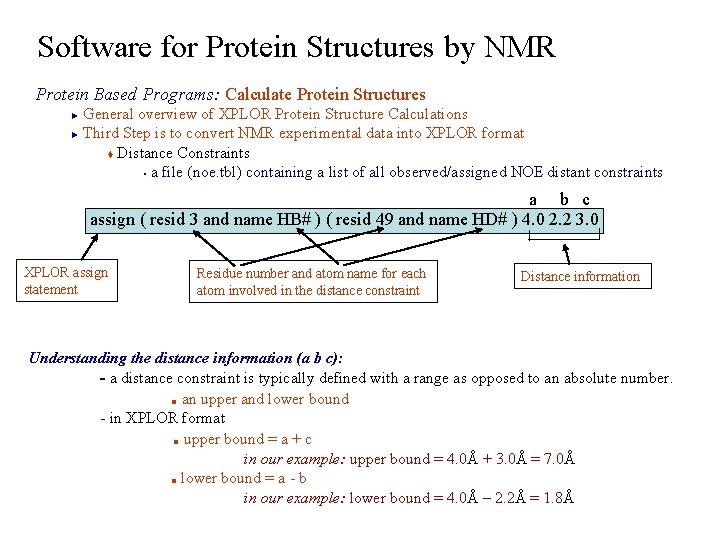
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations ► Third Step is to convert NMR experimental data into XPLOR format t Distance Constraints § a file (noe. tbl) containing a list of all observed/assigned NOE distant constraints ► a b c assign ( resid 3 and name HB# ) ( resid 49 and name HD# ) 4. 0 2. 2 3. 0 XPLOR assign statement Residue number and atom name for each atom involved in the distance constraint Distance information Understanding the distance information (a b c): - a distance constraint is typically defined with a range as opposed to an absolute number. ■ an upper and lower bound - in XPLOR format ■ upper bound = a + c in our example: upper bound = 4. 0Å + 3. 0Å = 7. 0Å ■ lower bound = a - b in our example: lower bound = 4. 0Å – 2. 2Å = 1. 8Å
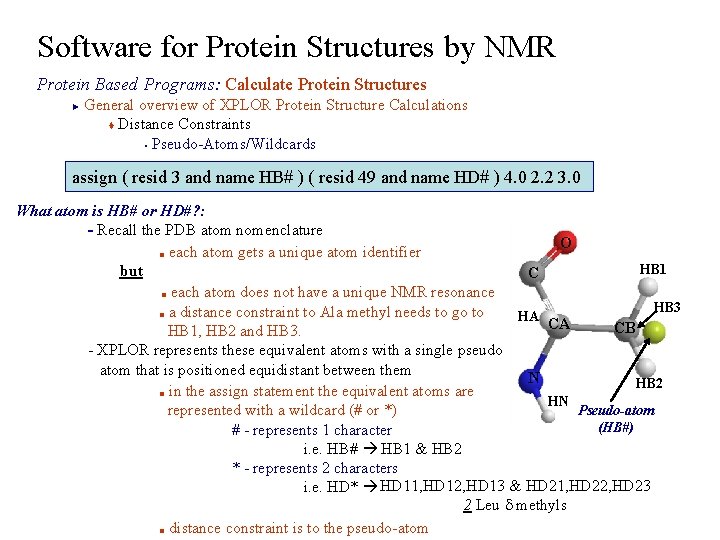
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations t Distance Constraints § Pseudo-Atoms/Wildcards ► assign ( resid 3 and name HB# ) ( resid 49 and name HD# ) 4. 0 2. 2 3. 0 What atom is HB# or HD#? : - Recall the PDB atom nomenclature O ■ each atom gets a unique atom identifier HB 1 but C ■ each atom does not have a unique NMR resonance HB 3 ■ a distance constraint to Ala methyl needs to go to HA CA CB HB 1, HB 2 and HB 3. - XPLOR represents these equivalent atoms with a single pseudo atom that is positioned equidistant between them N HB 2 ■ in the assign statement the equivalent atoms are HN Pseudo-atom represented with a wildcard (# or *) (HB#) # - represents 1 character i. e. HB# HB 1 & HB 2 * - represents 2 characters i. e. HD* HD 11, HD 12, HD 13 & HD 21, HD 22, HD 23 2 Leu d methyls ■ distance constraint is to the pseudo-atom
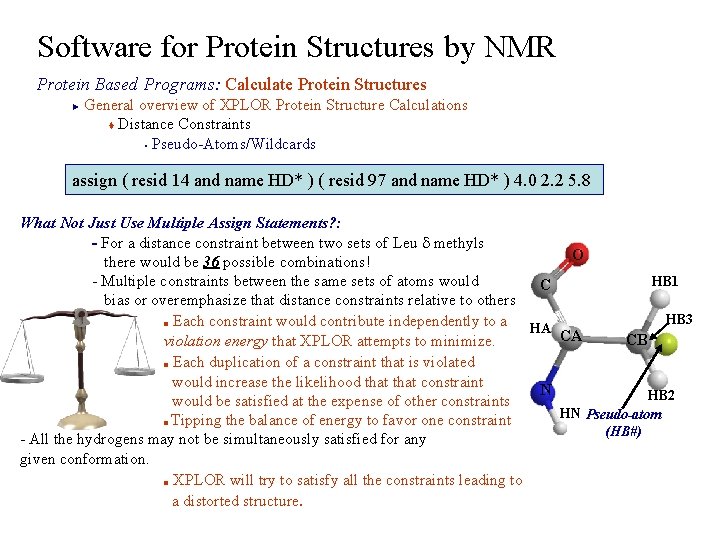
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations t Distance Constraints § Pseudo-Atoms/Wildcards ► assign ( resid 14 and name HD* ) ( resid 97 and name HD* ) 4. 0 2. 2 5. 8 What Not Just Use Multiple Assign Statements? : - For a distance constraint between two sets of Leu d methyls O there would be 36 possible combinations! HB 1 - Multiple constraints between the same sets of atoms would C bias or overemphasize that distance constraints relative to others HB 3 ■ Each constraint would contribute independently to a HA CA CB violation energy that XPLOR attempts to minimize. ■ Each duplication of a constraint that is violated would increase the likelihood that constraint N HB 2 would be satisfied at the expense of other constraints HN Pseudo-atom ■ Tipping the balance of energy to favor one constraint (HB#) - All the hydrogens may not be simultaneously satisfied for any given conformation. ■ XPLOR will try to satisfy all the constraints leading to a distorted structure.
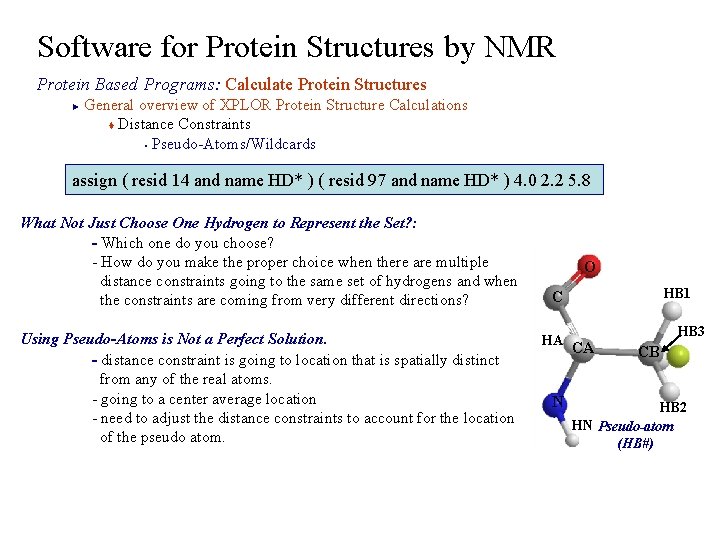
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations t Distance Constraints § Pseudo-Atoms/Wildcards ► assign ( resid 14 and name HD* ) ( resid 97 and name HD* ) 4. 0 2. 2 5. 8 What Not Just Choose One Hydrogen to Represent the Set? : - Which one do you choose? - How do you make the proper choice when there are multiple distance constraints going to the same set of hydrogens and when the constraints are coming from very different directions? O C HB 1 HB 3 Using Pseudo-Atoms is Not a Perfect Solution. HA CA CB - distance constraint is going to location that is spatially distinct from any of the real atoms. - going to a center average location N HB 2 - need to adjust the distance constraints to account for the location HN Pseudo-atom of the pseudo atom. (HB#)
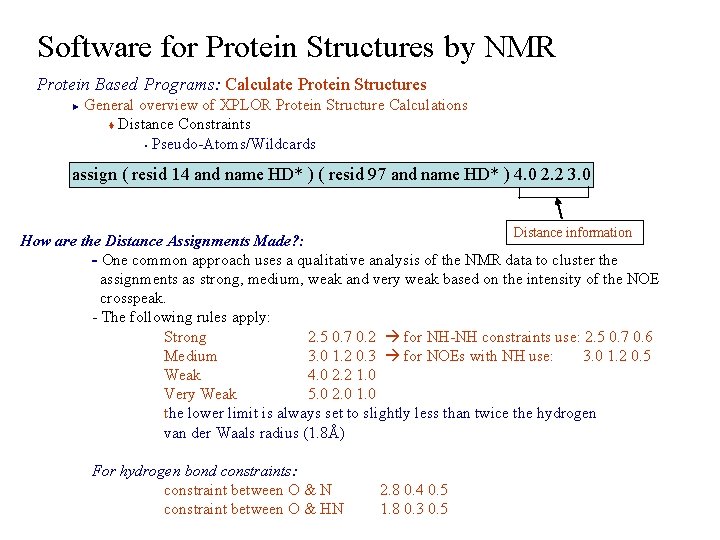
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations t Distance Constraints § Pseudo-Atoms/Wildcards ► assign ( resid 14 and name HD* ) ( resid 97 and name HD* ) 4. 0 2. 2 3. 0 Distance information How are the Distance Assignments Made? : - One common approach uses a qualitative analysis of the NMR data to cluster the assignments as strong, medium, weak and very weak based on the intensity of the NOE crosspeak. - The following rules apply: Strong 2. 5 0. 7 0. 2 for NH-NH constraints use: 2. 5 0. 7 0. 6 Medium 3. 0 1. 2 0. 3 for NOEs with NH use: 3. 0 1. 2 0. 5 Weak 4. 0 2. 2 1. 0 Very Weak 5. 0 2. 0 1. 0 the lower limit is always set to slightly less than twice the hydrogen van der Waals radius (1. 8Å) For hydrogen bond constraints: constraint between O & N constraint between O & HN 2. 8 0. 4 0. 5 1. 8 0. 3 0. 5
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations t Distance Constraints § Rules for pseudo-atom distance corrections: ► 1) For non stereoassigned Cb. H’s add 1. 0 to upper bound if HB# is used instead of HB 1 or HB 2 2) 1. 0 is added to upper bound for other methylenes if HG#, HD# or HE# is used instead of HG 1, HG 2, etc 3) 2. 0 is added to upper bound for aromatic Cd. H and Ce. H if HD# or HE# is used instead of HD 1, HD 2, etc for Tyr and Phe 4) 1. 5 is added to upper bound for CH 3 protons of methyls if HB# is used for Ala HB 1, HB 2, HB 3 or HG 2# is used for HG 21, HG 22, HG 23 for Thr or any other methyls 5) 2. 4 is added to upper bound for non stereoassigned methyls if HG* or HD* are used for Val or Leu methyls 6) Corrections are additive distance constraint from an HB# to a HD* would add 1. 0Å + 2. 4Å = 3. 4Å distance constraint between two HD* would add 2 x 2. 4Å= 4. 8Å
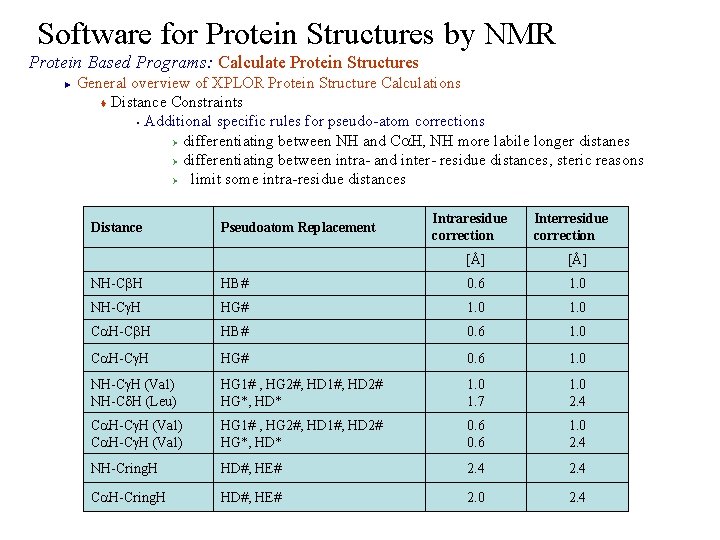
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations t Distance Constraints § Additional specific rules for pseudo-atom corrections Ø differentiating between NH and Ca. H, NH more labile longer distanes Ø differentiating between intra- and inter- residue distances, steric reasons Ø limit some intra-residue distances ► Intraresidue correction Interresidue correction Distance Pseudoatom Replacement [Å] NH-Cb. H HB# 0. 6 1. 0 NH-Cg. H HG# 1. 0 Ca. H-Cb. H HB# 0. 6 1. 0 Ca. H-Cg. H HG# 0. 6 1. 0 NH-Cg. H (Val) NH-Cd. H (Leu) HG 1# , HG 2#, HD 1#, HD 2# HG*, HD* 1. 0 1. 7 1. 0 2. 4 Ca. H-Cg. H (Val) HG 1# , HG 2#, HD 1#, HD 2# HG*, HD* 0. 6 1. 0 2. 4 NH-Cring. H HD#, HE# 2. 4 Ca. H-Cring. H HD#, HE# 2. 0 2. 4
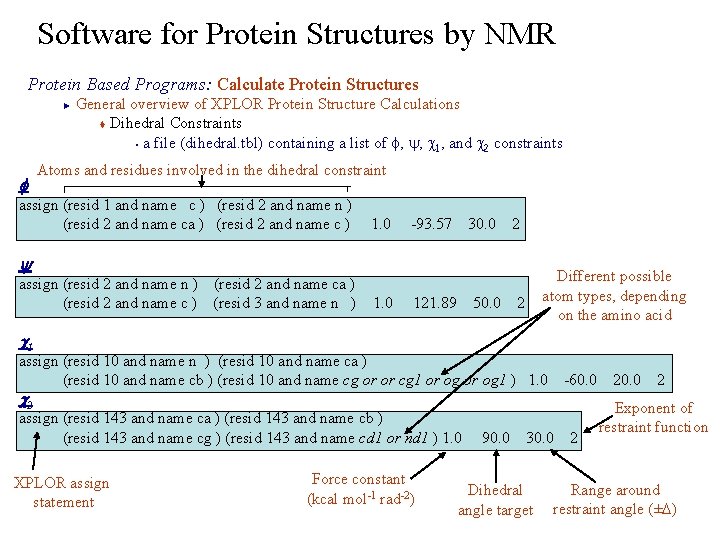
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations t Dihedral Constraints § a file (dihedral. tbl) containing a list of f, y, c 1, and c 2 constraints ► f Atoms and residues involved in the dihedral constraint assign (resid 1 and name c ) (resid 2 and name n ) (resid 2 and name ca ) (resid 2 and name c ) 1. 0 -93. 57 30. 0 2 y assign (resid 2 and name n ) (resid 2 and name ca ) (resid 2 and name c ) (resid 3 and name n ) 1. 0 121. 89 50. 0 2 Different possible atom types, depending on the amino acid c 1 assign (resid 10 and name n ) (resid 10 and name ca ) (resid 10 and name cb ) (resid 10 and name cg or or cg 1 or og 1 ) 1. 0 -60. 0 2 c 2 assign (resid 143 and name ca ) (resid 143 and name cb ) (resid 143 and name cg ) (resid 143 and name cd 1 or nd 1 ) 1. 0 90. 0 30. 0 2 XPLOR assign statement Force constant (kcal mol-1 rad-2) Dihedral angle target Exponent of restraint function Range around restraint angle (±D)
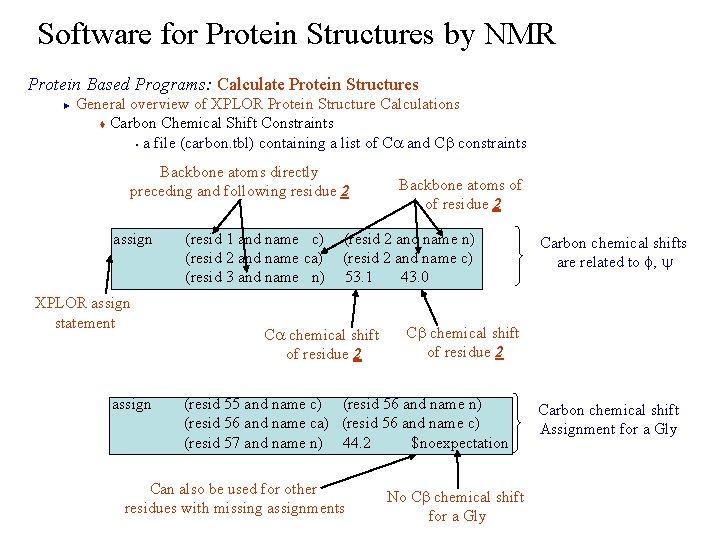
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations t Carbon Chemical Shift Constraints § a file (carbon. tbl) containing a list of Ca and Cb constraints ► Backbone atoms directly preceding and following residue 2 assign XPLOR assign statement assign Backbone atoms of of residue 2 (resid 1 and name c) (resid 2 and name n) (resid 2 and name ca) (resid 2 and name c) (resid 3 and name n) 53. 1 43. 0 Ca chemical shift of residue 2 Cb chemical shift of residue 2 (resid 55 and name c) (resid 56 and name n) (resid 56 and name ca) (resid 56 and name c) (resid 57 and name n) 44. 2 $noexpectation Can also be used for other residues with missing assignments Carbon chemical shifts are related to f, y No Cb chemical shift for a Gly Carbon chemical shift Assignment for a Gly
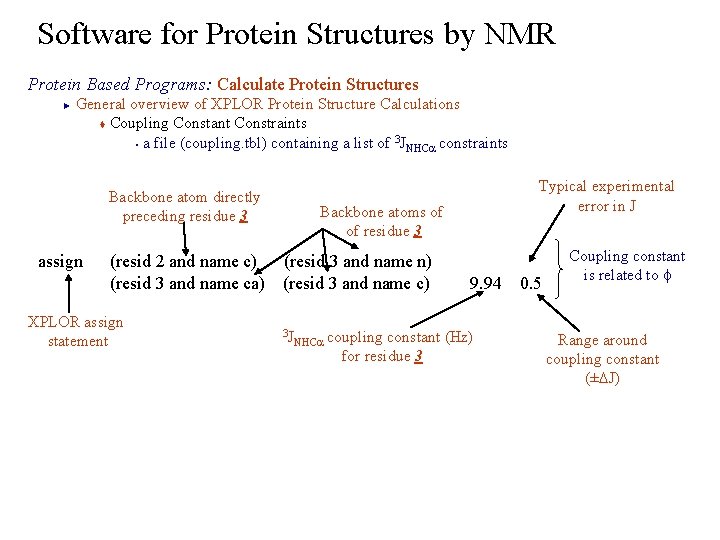
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations t Coupling Constant Constraints 3 § a file (coupling. tbl) containing a list of JNHCa constraints ► Backbone atom directly preceding residue 3 assign Backbone atoms of of residue 3 (resid 2 and name c) (resid 3 and name n) (resid 3 and name ca) (resid 3 and name c) XPLOR assign statement Typical experimental error in J 3 J 9. 94 0. 5 NHCa coupling constant (Hz) for residue 3 Coupling constant is related to f Range around coupling constant (±DJ)
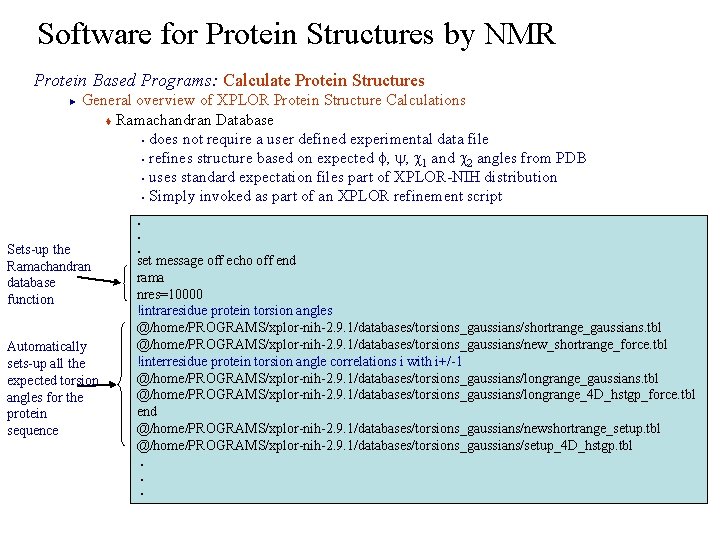
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations t Ramachandran Database § does not require a user defined experimental data file § refines structure based on expected f, y, c 1 and c 2 angles from PDB § uses standard expectation files part of XPLOR-NIH distribution § Simply invoked as part of an XPLOR refinement script ► Sets-up the Ramachandran database function Automatically sets-up all the expected torsion angles for the protein sequence . . . set message off echo off end rama nres=10000 !intraresidue protein torsion angles @/home/PROGRAMS/xplor-nih-2. 9. 1/databases/torsions_gaussians/shortrange_gaussians. tbl @/home/PROGRAMS/xplor-nih-2. 9. 1/databases/torsions_gaussians/new_shortrange_force. tbl !interresidue protein torsion angle correlations i with i+/-1 @/home/PROGRAMS/xplor-nih-2. 9. 1/databases/torsions_gaussians/longrange_gaussians. tbl @/home/PROGRAMS/xplor-nih-2. 9. 1/databases/torsions_gaussians/longrange_4 D_hstgp_force. tbl end @/home/PROGRAMS/xplor-nih-2. 9. 1/databases/torsions_gaussians/newshortrange_setup. tbl @/home/PROGRAMS/xplor-nih-2. 9. 1/databases/torsions_gaussians/setup_4 D_hstgp. tbl. . .
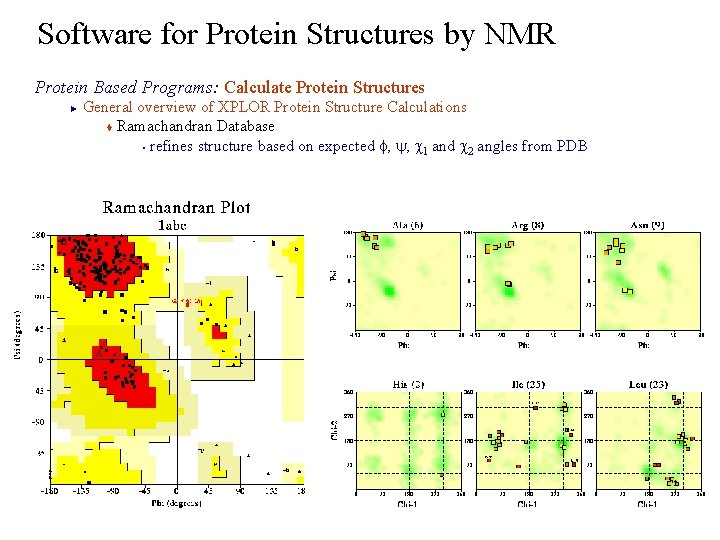
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations t Ramachandran Database § refines structure based on expected f, y, c 1 and c 2 angles from PDB ►
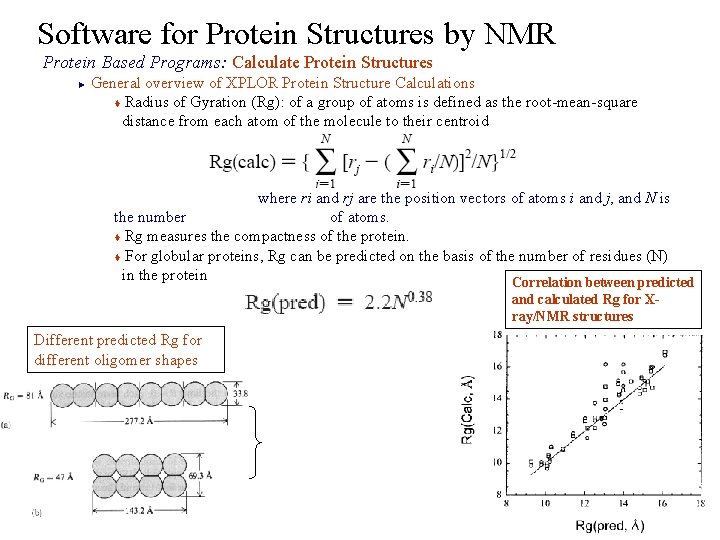
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations t Radius of Gyration (Rg): of a group of atoms is defined as the root-mean-square distance from each atom of the molecule to their centroid ► where ri and rj are the position vectors of atoms i and j, and N is the number of atoms. t Rg measures the compactness of the protein. t For globular proteins, Rg can be predicted on the basis of the number of residues (N) in the protein Correlation between predicted and calculated Rg for Xray/NMR structures Different predicted Rg for different oligomer shapes
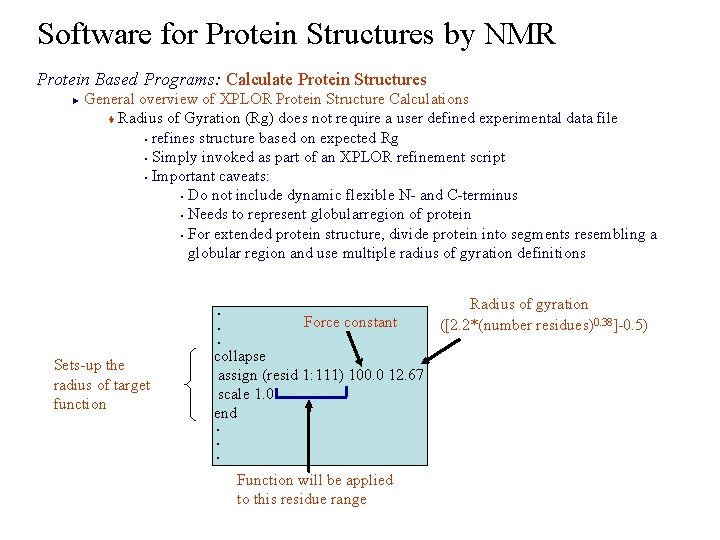
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations t Radius of Gyration (Rg) does not require a user defined experimental data file § refines structure based on expected Rg § Simply invoked as part of an XPLOR refinement script § Important caveats: § Do not include dynamic flexible N- and C-terminus § Needs to represent globularregion of protein § For extended protein structure, divide protein into segments resembling a globular region and use multiple radius of gyration definitions ► . . . Sets-up the radius of target function Force constant collapse assign (resid 1: 111) 100. 0 12. 67 scale 1. 0 end. . . Function will be applied to this residue range Radius of gyration ([2. 2*(number residues)0. 38]-0. 5)
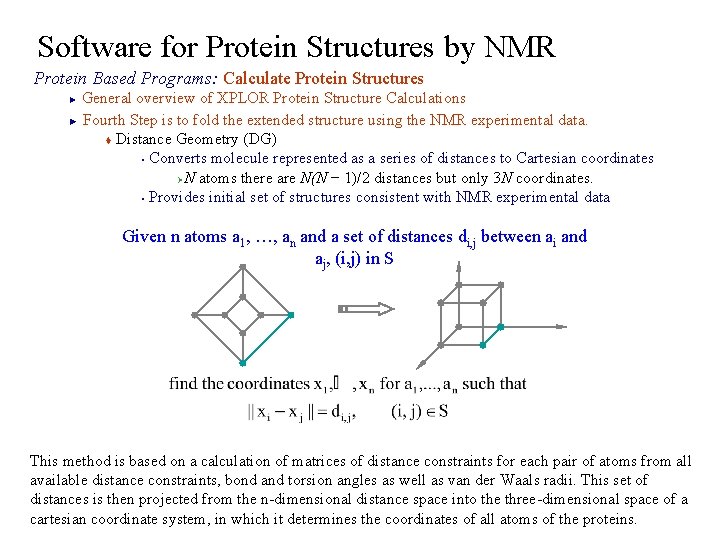
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations ► Fourth Step is to fold the extended structure using the NMR experimental data. t Distance Geometry (DG) § Converts molecule represented as a series of distances to Cartesian coordinates ØN atoms there are N(N − 1)/2 distances but only 3 N coordinates. § Provides initial set of structures consistent with NMR experimental data ► Given n atoms a 1, …, an and a set of distances di, j between ai and aj, (i, j) in S This method is based on a calculation of matrices of distance constraints for each pair of atoms from all available distance constraints, bond and torsion angles as well as van der Waals radii. This set of distances is then projected from the n-dimensional distance space into the three-dimensional space of a cartesian coordinate system, in which it determines the coordinates of all atoms of the proteins.
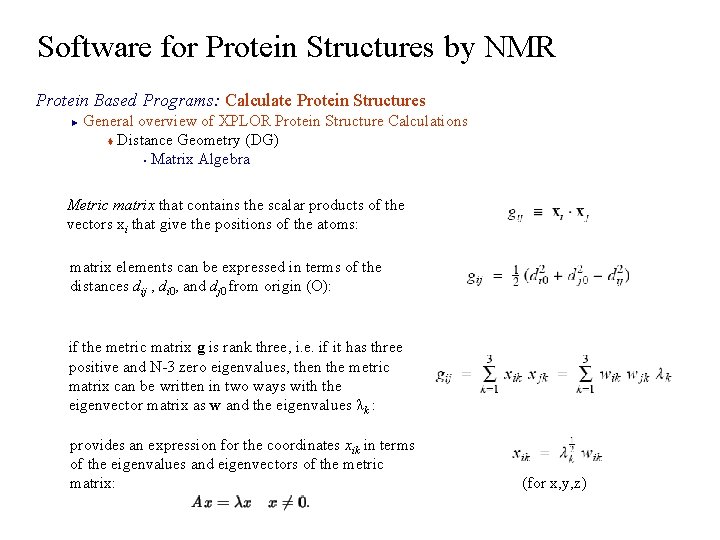
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations t Distance Geometry (DG) § Matrix Algebra ► Metric matrix that contains the scalar products of the vectors xi that give the positions of the atoms: matrix elements can be expressed in terms of the distances dij , di 0, and dj 0 from origin (O): if the metric matrix g is rank three, i. e. if it has three positive and N-3 zero eigenvalues, then the metric matrix can be written in two ways with the eigenvector matrix as w and the eigenvalues λk : provides an expression for the coordinates xik in terms of the eigenvalues and eigenvectors of the metric matrix: (for x, y, z)
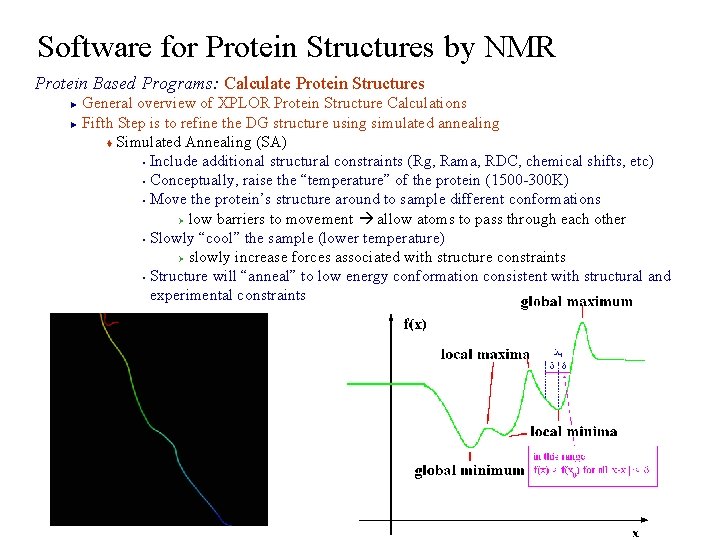
Software for Protein Structures by NMR Protein Based Programs: Calculate Protein Structures General overview of XPLOR Protein Structure Calculations ► Fifth Step is to refine the DG structure using simulated annealing t Simulated Annealing (SA) § Include additional structural constraints (Rg, Rama, RDC, chemical shifts, etc) § Conceptually, raise the “temperature” of the protein (1500 -300 K) § Move the protein’s structure around to sample different conformations Ø low barriers to movement allow atoms to pass through each other § Slowly “cool” the sample (lower temperature) Ø slowly increase forces associated with structure constraints § Structure will “anneal” to low energy conformation consistent with structural and experimental constraints ►
- Slides: 55