small RNAs Small molecules big functions Brief history

small. RNAs Small molecules, big functions

Brief history • The first described micro. RNA, lin-4 was cloned and characterised as a translational repressor of developmental timing from Caenorhabditis. elegans by Lee et al (1993) and Wightman et al (1993). • The transcript of this gene was highly unusual as it was non-coding, and produced extremely small transcripts (22 nt) from hairpin structured RNA precursors. • Second micro. RNA, let-7 was also cloned from C. elegans (Reinhart et al, 2000). • There are currently 474 human cloned and characterised micro. RNA sequences deposited in the mi. RBase database (http: //microrna. sanger. ac. uk/sequences/) • Micro. RNAs primarily function as translational repressors by binding to complementary target sequences in the 3’ UTR (untranslated region) of m. RNA.

Brief history • Between 10– 30% of all human genes are a target for micro. RNA regulation (John et al, 2004; Lewis et al, 2005). • A single target gene often contains putative binding sites for multiple micro. RNAs that can bind cooperatively , allowing micro. RNAs to form complex regulatory control networks. • micro. RNAs play key regulatory roles in control of haematopoiesis, developmental timing, cell differentiation, apoptosis, cell proliferation and organ development as well as in cancer, infectious disease, genetic disorders (Lin et al, 2006) and even heart disease (van Rooij et al, 2006).
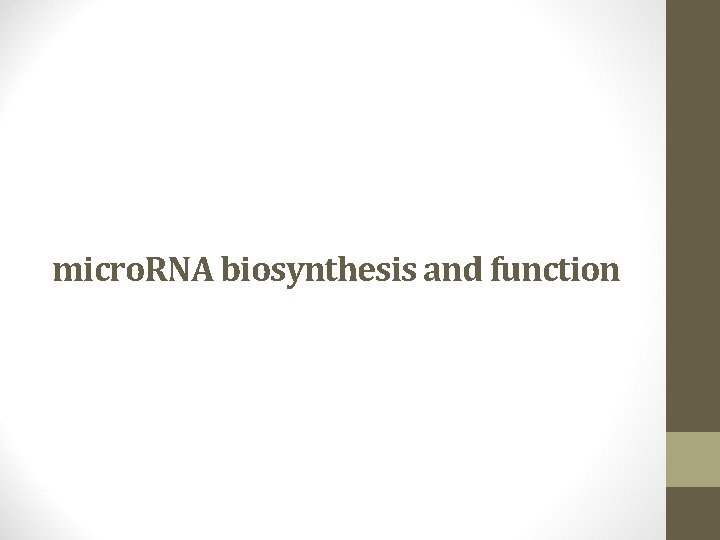
micro. RNA biosynthesis and function

• Micro. RNAs are transcribed in a RNA Polymerase II-dependent manner as large polyadenylated pri-micro. RNAs. • RNAPII catalyzes the transcription of DNA to synthesize precursors of m. RNA and most sn. RNA and micro. RNA Yang CGFR 16: 397, 2005
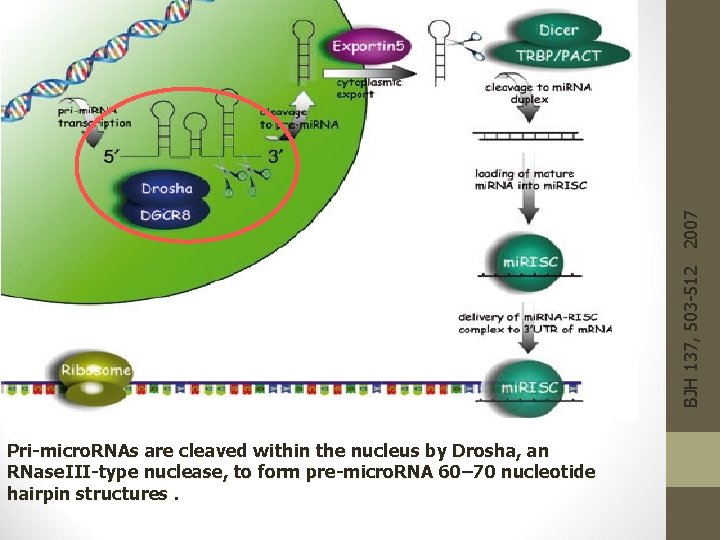
BJH 137, 503 -512 2007 Pri-micro. RNAs are cleaved within the nucleus by Drosha, an RNase. III-type nuclease, to form pre-micro. RNA 60– 70 nucleotide hairpin structures.
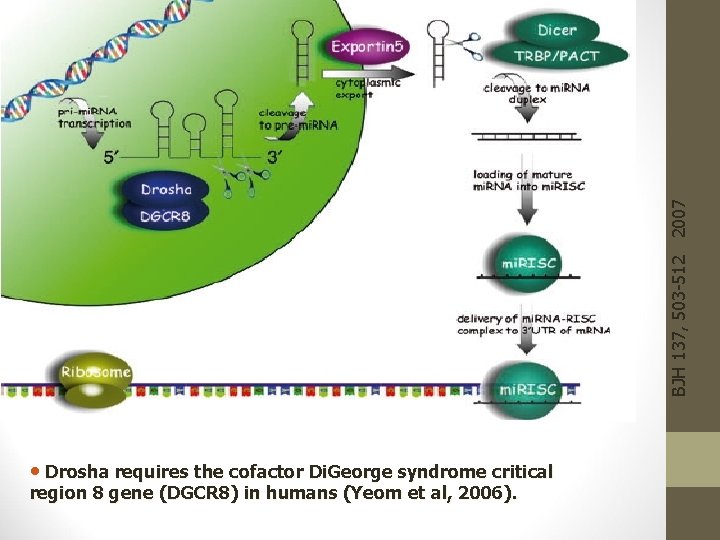
BJH 137, 503 -512 2007 • Drosha requires the cofactor Di. George syndrome critical region 8 gene (DGCR 8) in humans (Yeom et al, 2006).
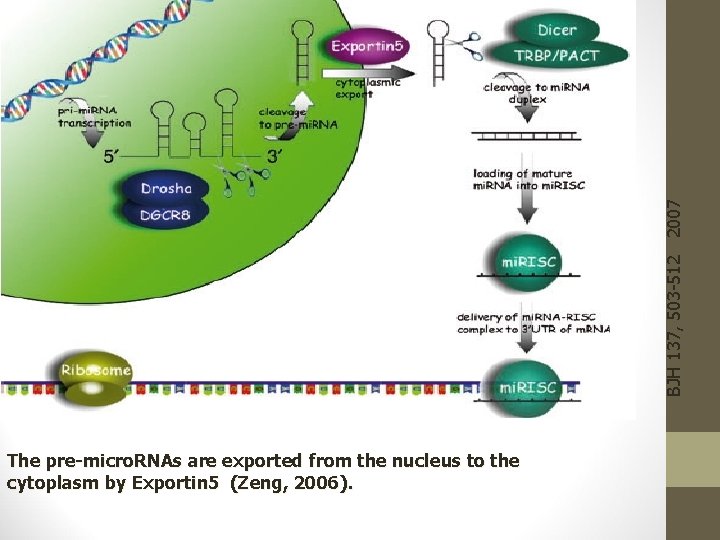
BJH 137, 503 -512 2007 The pre-micro. RNAs are exported from the nucleus to the cytoplasm by Exportin 5 (Zeng, 2006).
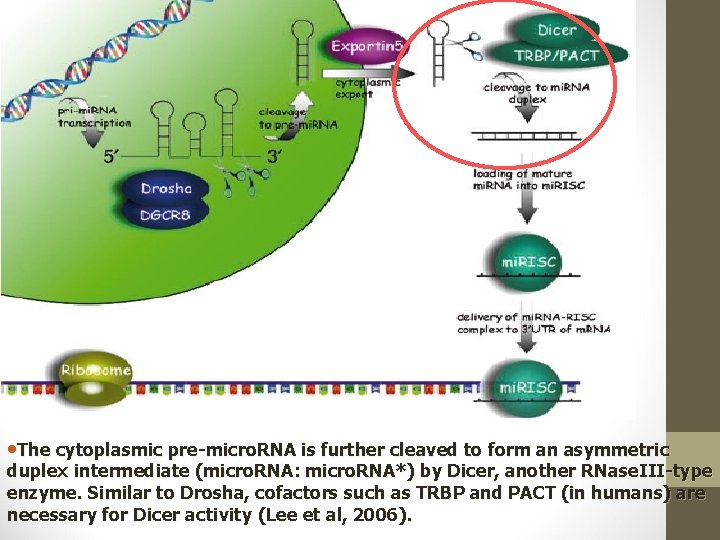
• The cytoplasmic pre-micro. RNA is further cleaved to form an asymmetric duplex intermediate (micro. RNA: micro. RNA*) by Dicer, another RNase. III-type enzyme. Similar to Drosha, cofactors such as TRBP and PACT (in humans) are necessary for Dicer activity (Lee et al, 2006).
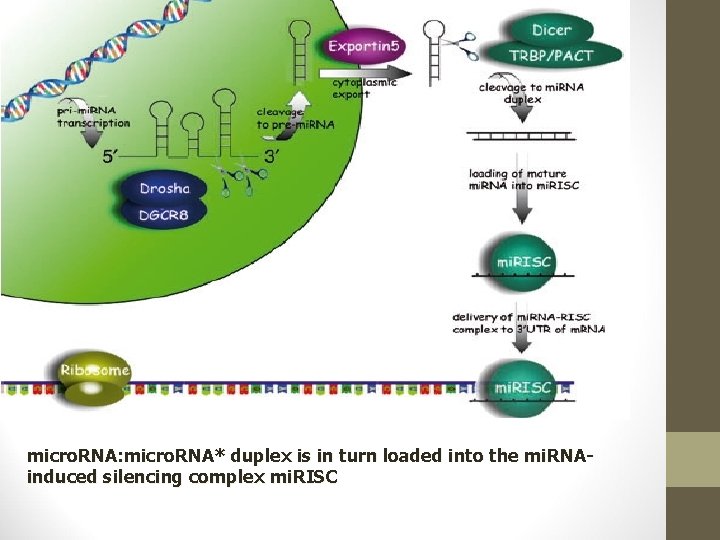
micro. RNA: micro. RNA* duplex is in turn loaded into the mi. RNAinduced silencing complex mi. RISC
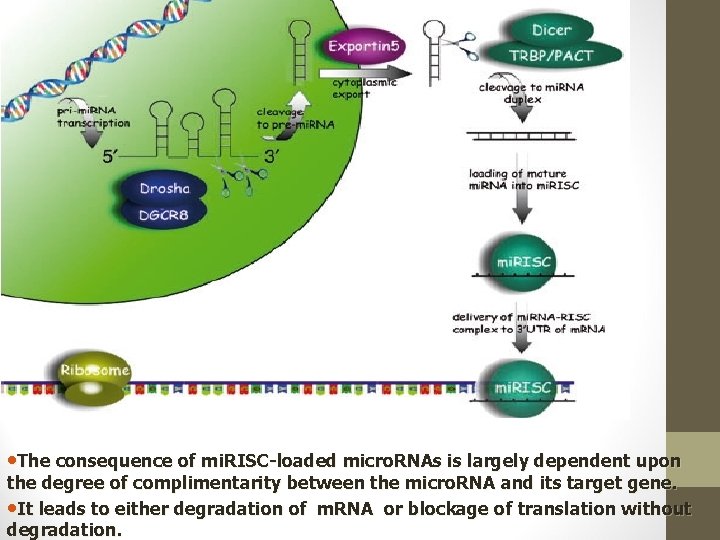
• The consequence of mi. RISC-loaded micro. RNAs is largely dependent upon the degree of complimentarity between the micro. RNA and its target gene. • It leads to either degradation of m. RNA or blockage of translation without degradation.

The choice of posttranscriptional mechanisms is not determined by whether the small silencing RNA originated an si. RNA or a mi. RNA but instead is determined by the identity of the target. Cell, Vol. 116, 281– 297, January 23, 2004

Aberrant expression of micro. RNA • The majority of human micro. RNAs are located at cancer-associated genomic regions (Calin et al, 2004 a). • micro. RNA expression profiling can distinguish cancers according to diagnosis and developmental stage of the tumour to a greater degree of accuracy than traditional gene expression analysis (Lu et al, 2005). • Micro. RNAs play a direct role in oncogenesis as they can function as both oncogenes (e. g. MIRN 155 and members of MIRN 17– 92 cluster) and tumour suppressor molecules [e. g. MIRN 15 A (mi. R-15 a) and MIRN 16 -1 (mi. R-16 -1)]. • Aberrant expression of specific micro. RNAs has now been associated with many types of cancer including solid and haematopoietic tumours.

micro. RNA expression in leukaemia • Expression levels of MIRN 15 A and MIRN 16 -1, encoded within the 13 q 14 region, were downregulated in 75% of CLL cases that harboured this chromosomal abnormality. • These micro. RNAs were subsequently shown to target BCL 2 and to induce apoptosis in vitro, suggesting they have tumour-suppressor role in CLL (Cimmino et al, 2005). • MIRN 16 -1 negatively regulates cellular growth and cell cycle progression (Linsley et al, 2007). • A follow-up study (Calin et al, 2005) of 94 CLL cases, defined a prognostically significant 13 -gene micro. RNA signature by expression profiling. • Moreover two of the CLL patients were found to have germline mutations in the MIRN 16 -1/MIRN 15 A precursor sequence that resulted in reduced expression levels of these micro. RNAs both in vitro and in vivo.

RNAi gene therapy application • Viral infections: - HIV - Hep B - Hep C - RSV • Cancer • Neurodegenerative disorders: - Spinocerebellar Ataxias - Huntington disease - alzheimer disease • Ocular disorders (Macular degeneration) • Stem cell biology and therapy
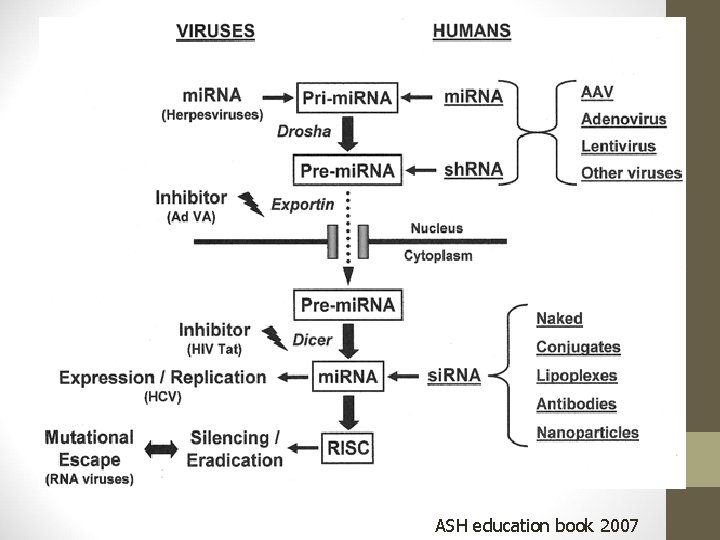
ASH education book 2007
http: //www. dharmacon. com/designcenterpage. aspx
http: //www. mirbase. org/index. shtml
http: //diana. cslab. ece. ntua. gr/? sec=home

- Slides: 24