Singlemolecule detection of DNA transcription and replication Transcription

- Slides: 34

Single-molecule detection of DNA transcription and replication

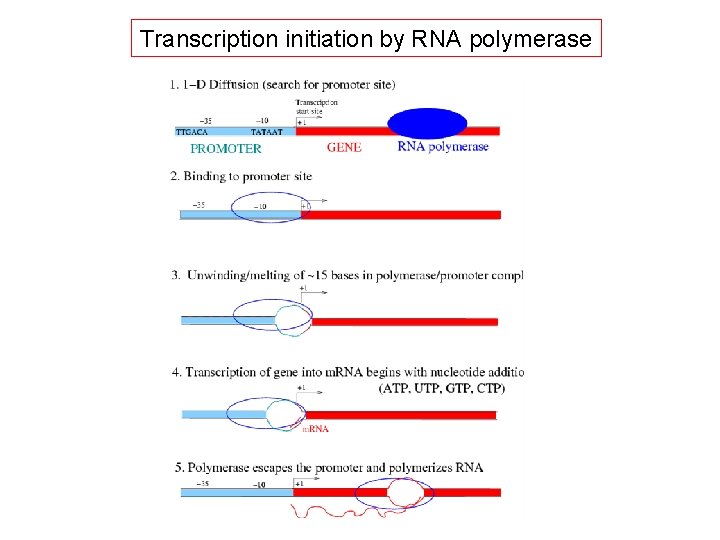
Transcription initiation by RNA polymerase

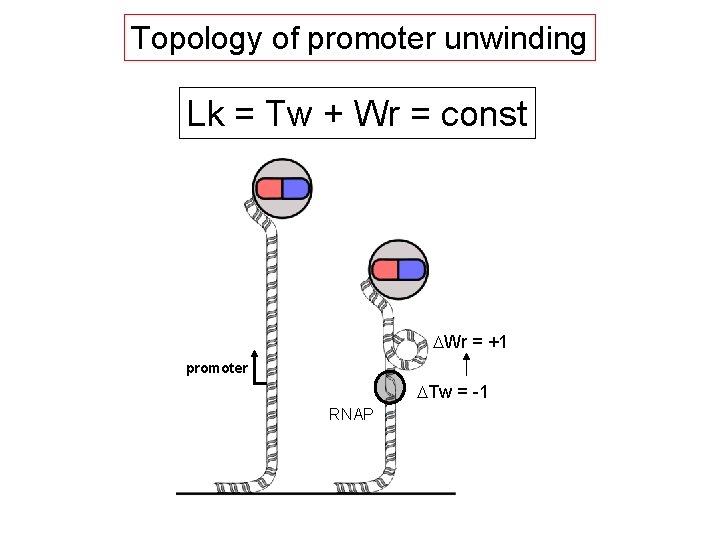
Topology of promoter unwinding Lk = Tw + Wr = const DWr = +1 promoter DTw = -1 RNAP

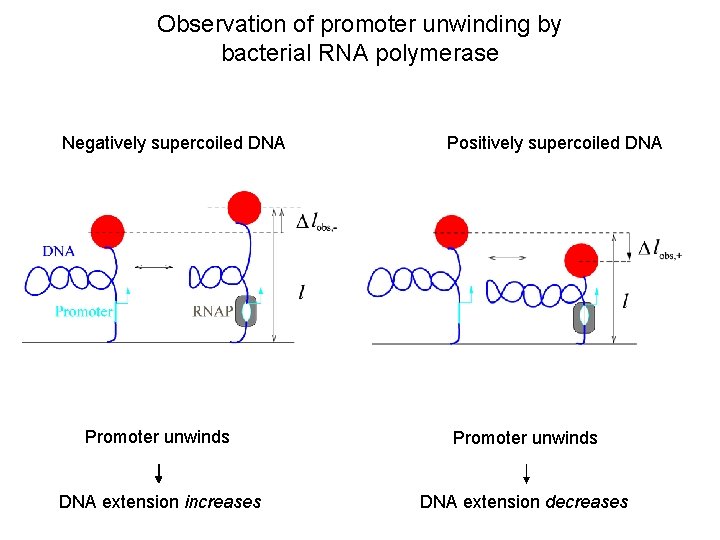
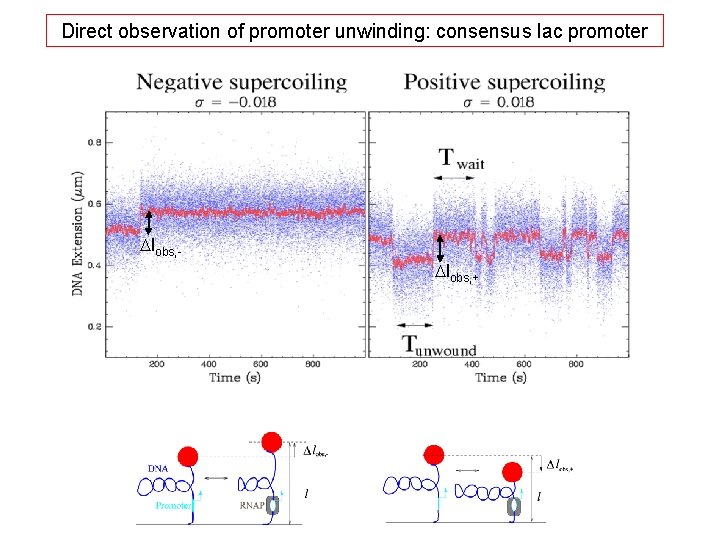
Observation of promoter unwinding by bacterial RNA polymerase Negatively supercoiled DNA Positively supercoiled DNA Promoter unwinds DNA extension increases DNA extension decreases

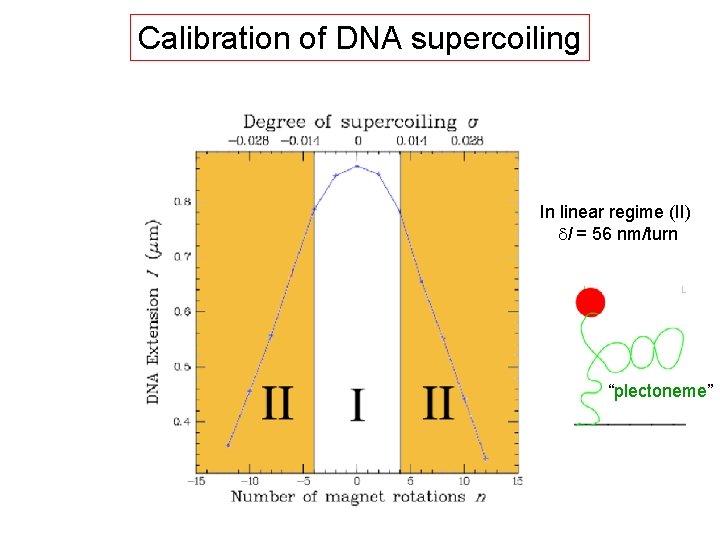
Calibration of DNA supercoiling In linear regime (II) dl = 56 nm/turn “plectoneme”

Direct observation of promoter unwinding: consensus lac promoter Dlobs, +

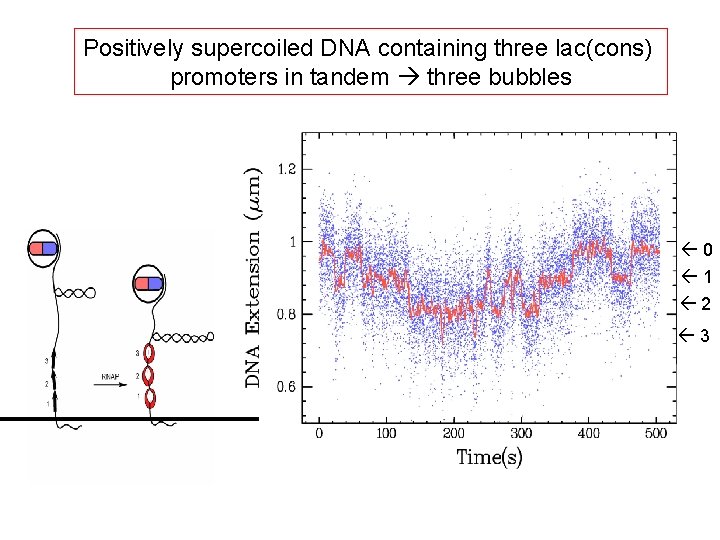
Positively supercoiled DNA containing three lac(cons) promoters in tandem three bubbles 0 1 2 3

More Control Experiments 1. No unwinding is observed with a DNA template having no promoter; 2. No promoter unwinding is observed in the absence of the initiation factor s; 3. No unwinding is observed at temperatures below 23 C; 4. Unwinding is abolished by prior addition of heparin (binds free RNAP);

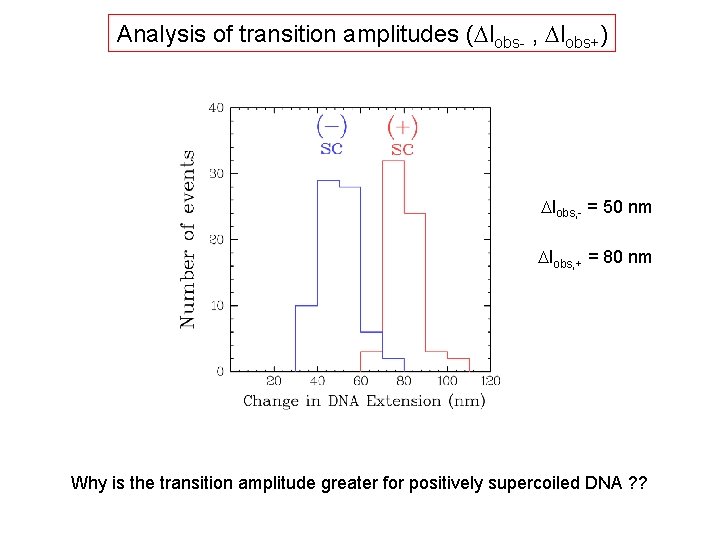
Analysis of transition amplitudes (Dlobs- , Dlobs+) Dlobs, - = 50 nm Dlobs, + = 80 nm Why is the transition amplitude greater for positively supercoiled DNA ? ?

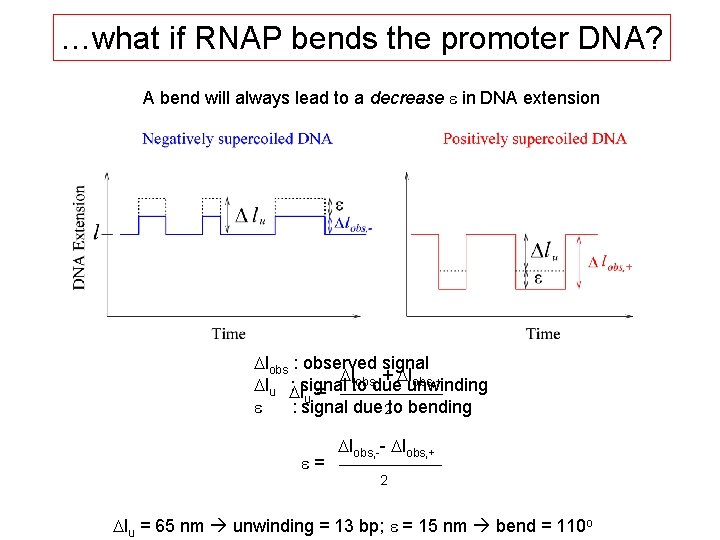
…what if RNAP bends the promoter DNA? A bend will always lead to a decrease e in DNA extension Dlobs : observed signal Dlobs, -+ Dlobs, + Dlu Dl : signal to due unwinding u = e : signal due 2 to bending e= Dlobs, -- Dlobs, + 2 Dlu = 65 nm unwinding = 13 bp; e = 15 nm bend = 110 o

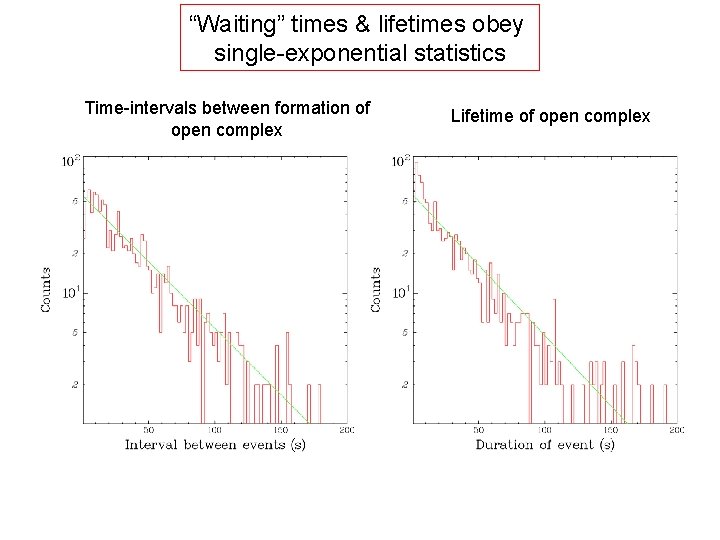
“Waiting” times & lifetimes obey single-exponential statistics Time-intervals between formation of open complex Lifetime of open complex

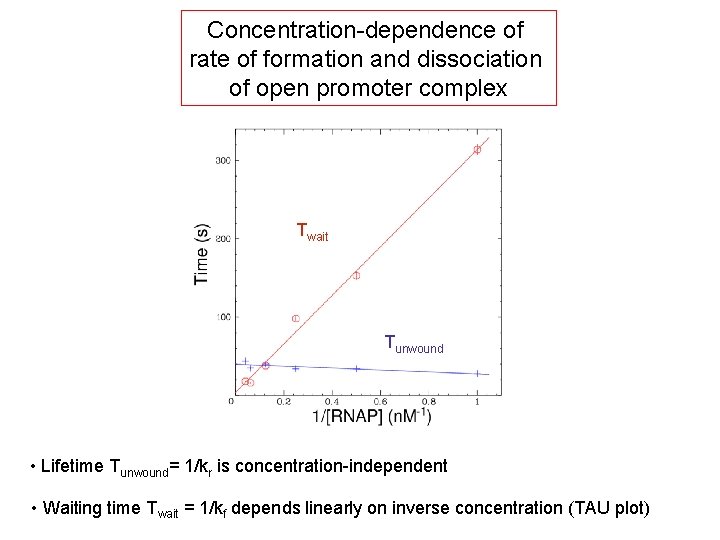
Concentration-dependence of rate of formation and dissociation of open promoter complex Twait Tunwound • Lifetime Tunwound= 1/kr is concentration-independent • Waiting time Twait = 1/kf depends linearly on inverse concentration (TAU plot)

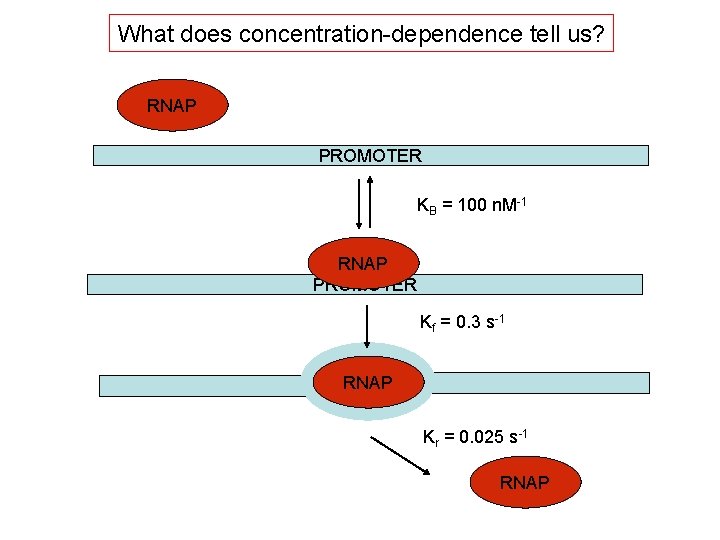
What does concentration-dependence tell us? RNAP PROMOTER KB = 100 n. M-1 RNAP PROMOTER Kf = 0. 3 s-1 RNAP Kr = 0. 025 s-1 RNAP

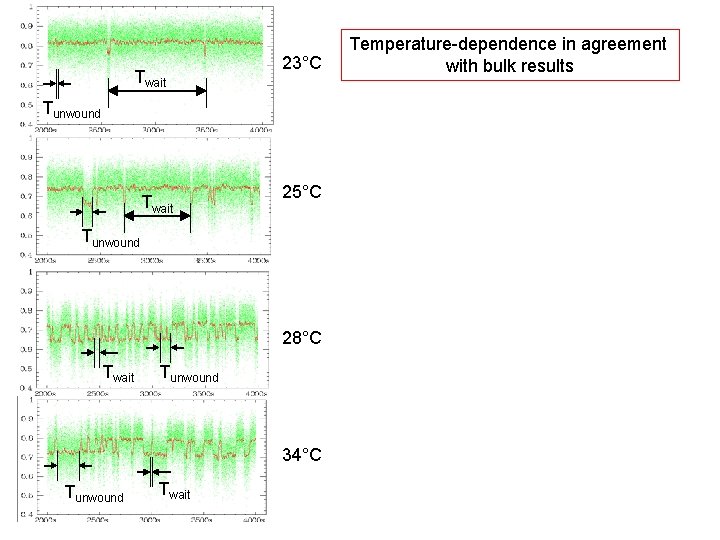
Twait 23°C Tunwound Twait 25°C Tunwound 28°C Twait Tunwound 34°C Tunwound Twait Temperature-dependence in agreement with bulk results

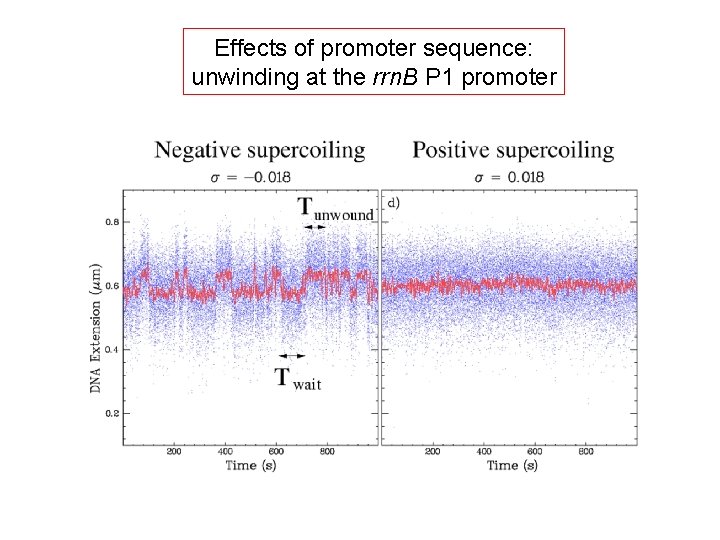
Effects of promoter sequence: unwinding at the rrn. B P 1 promoter

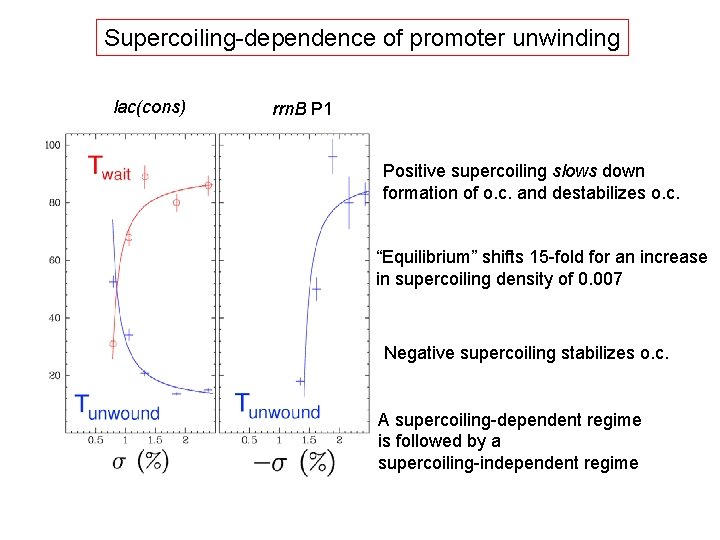
Supercoiling-dependence of promoter unwinding lac(cons) rrn. B P 1 Positive supercoiling slows down formation of o. c. and destabilizes o. c. “Equilibrium” shifts 15 -fold for an increase in supercoiling density of 0. 007 Negative supercoiling stabilizes o. c. A supercoiling-dependent regime is followed by a supercoiling-independent regime

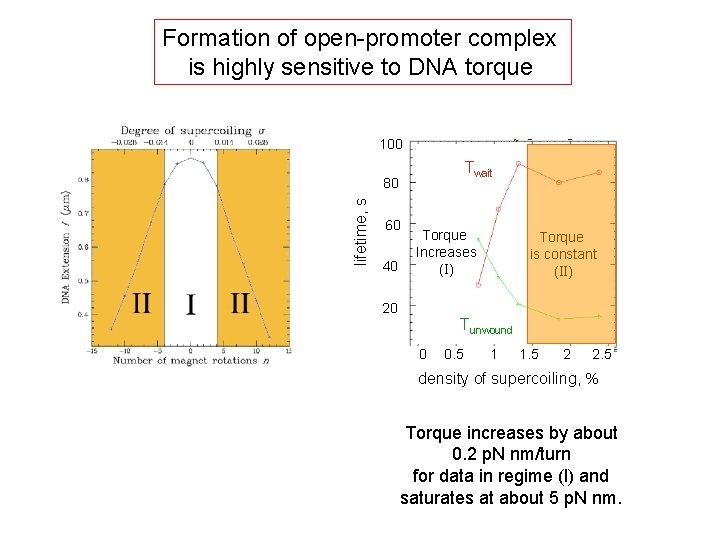
Formation of open-promoter complex is highly sensitive to DNA torque 100 Twait lifetime, s 80 60 40 Torque Increases (I) 20 Torque is constant (II) Tunwound 0 0. 5 1 1. 5 2 2. 5 density of supercoiling, % Torque increases by about 0. 2 p. N nm/turn for data in regime (I) and saturates at about 5 p. N nm.

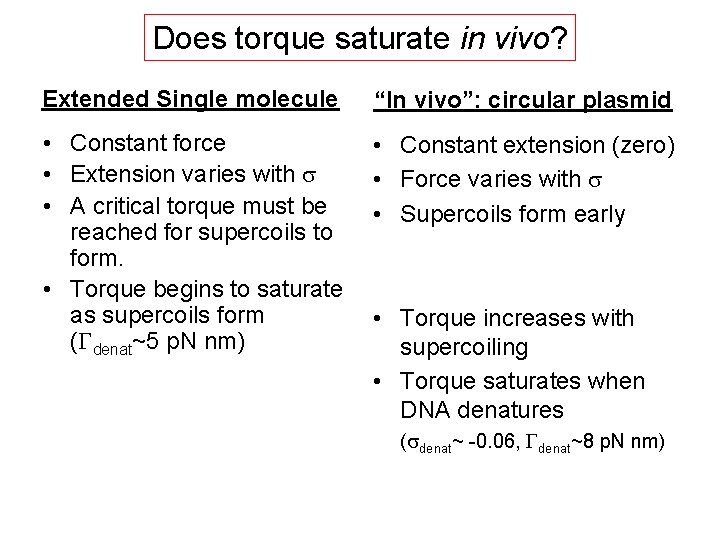
Does torque saturate in vivo? Extended Single molecule “In vivo”: circular plasmid • Constant force • Extension varies with s • A critical torque must be reached for supercoils to form. • Torque begins to saturate as supercoils form (Gdenat~5 p. N nm) • Constant extension (zero) • Force varies with s • Supercoils form early • Torque increases with supercoiling • Torque saturates when DNA denatures (sdenat~ -0. 06, Gdenat~8 p. N nm)

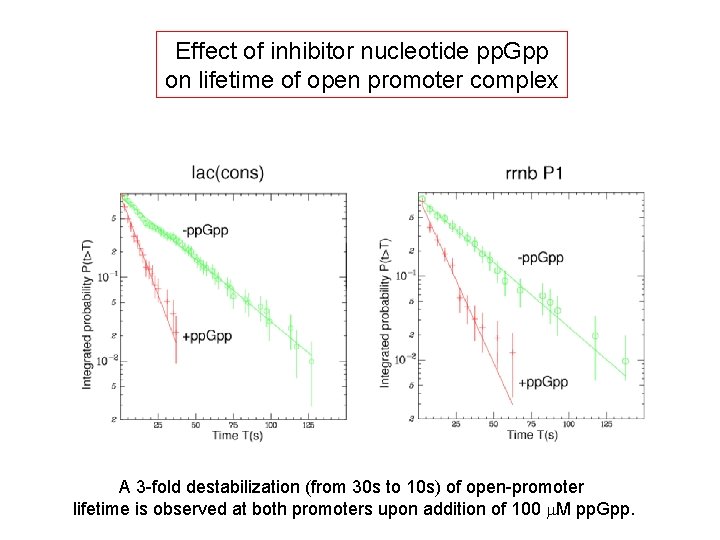
Effect of inhibitor nucleotide pp. Gpp on lifetime of open promoter complex A 3 -fold destabilization (from 30 s to 10 s) of open-promoter lifetime is observed at both promoters upon addition of 100 m. M pp. Gpp.

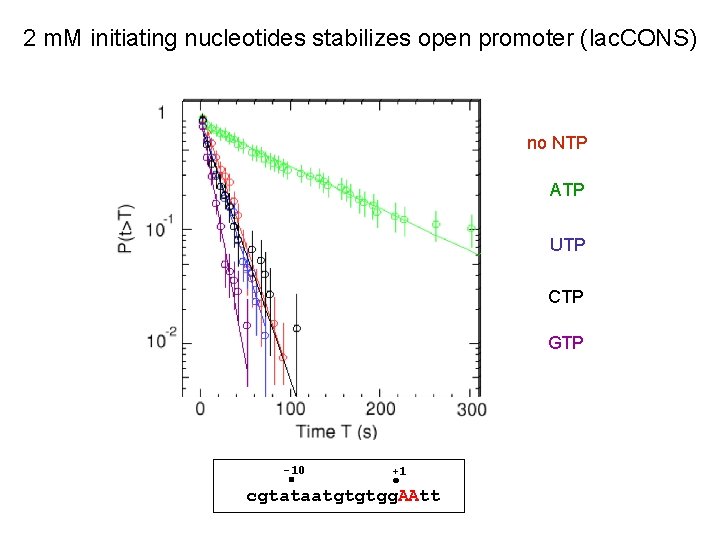
2 m. M initiating nucleotides stabilizes open promoter (lac. CONS) no NTP ATP UTP CTP GTP -10 +1 cgtataatgtgtgg. AAtt

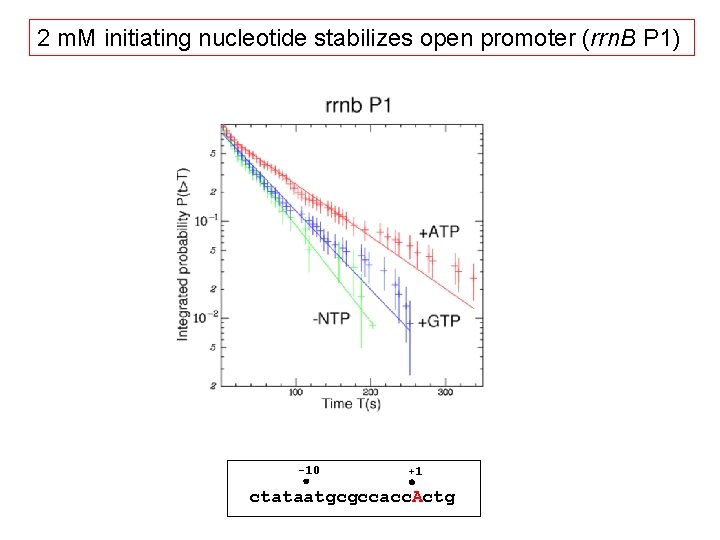
2 m. M initiating nucleotide stabilizes open promoter (rrn. B P 1) -10 +1 ctataatgcgccacc. Actg

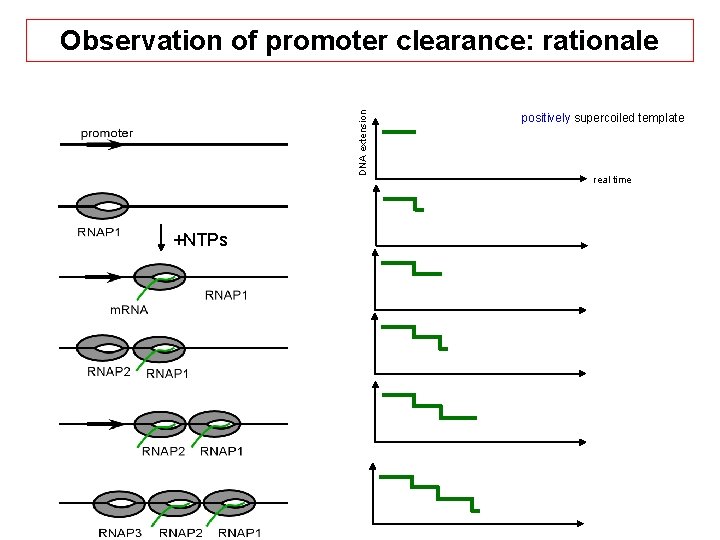
DNA extension Observation of promoter clearance: rationale +NTPs positively supercoiled template real time

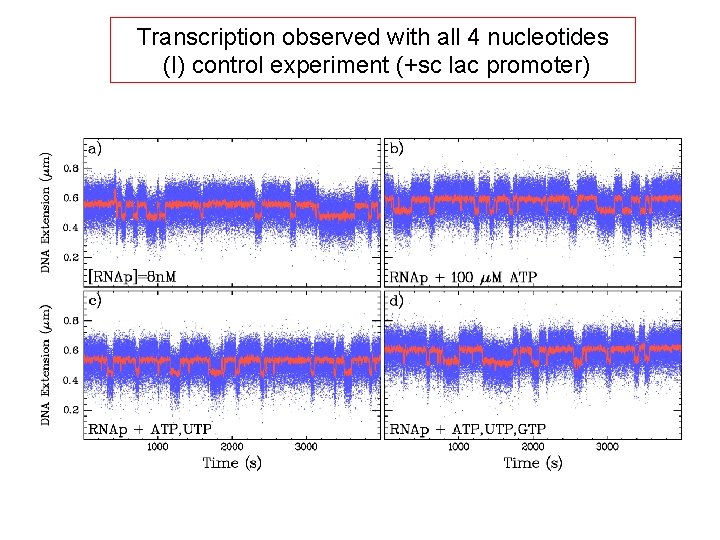
Transcription observed with all 4 nucleotides (I) control experiment (+sc lac promoter)

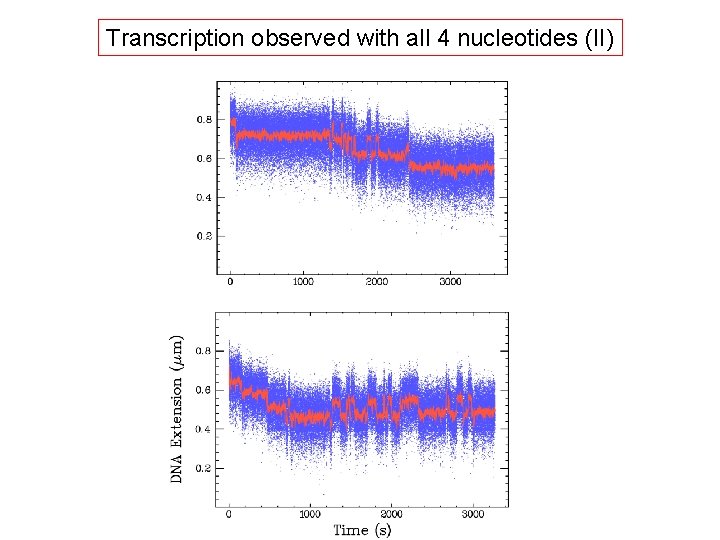
Transcription observed with all 4 nucleotides (II)

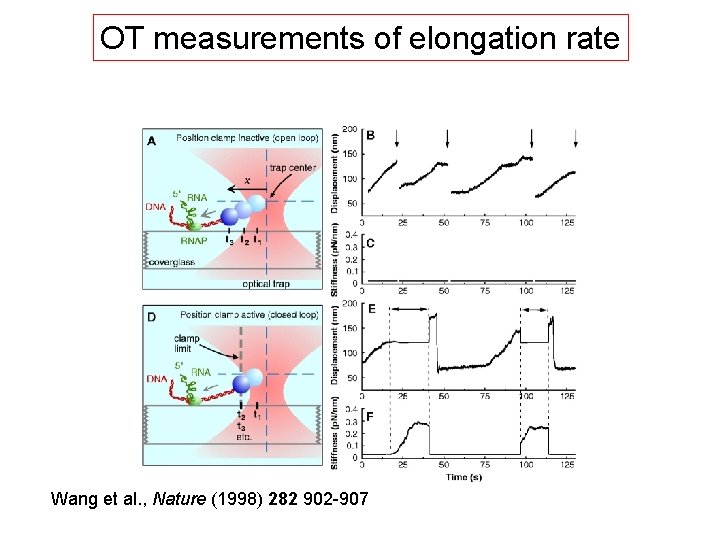
OT measurements of elongation rate Wang et al. , Nature (1998) 282 902 -907

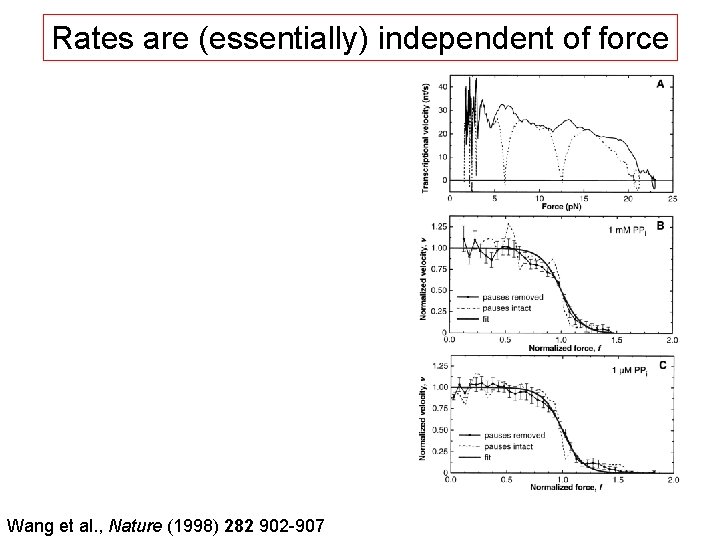
Rates are (essentially) independent of force Wang et al. , Nature (1998) 282 902 -907

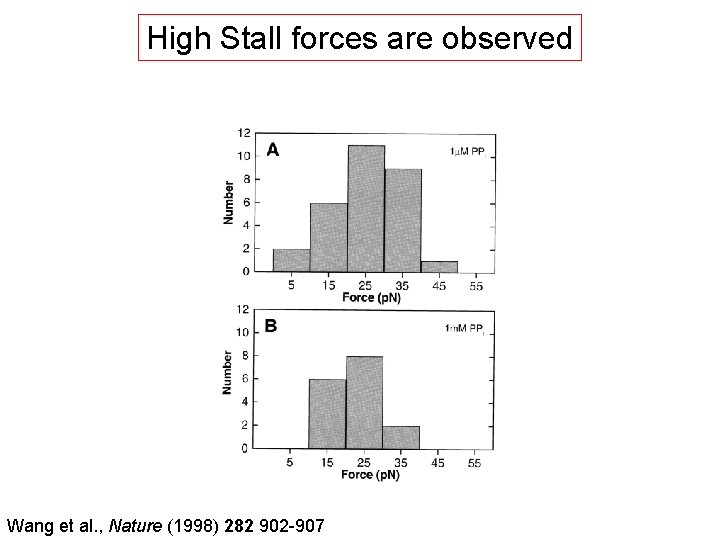
High Stall forces are observed Wang et al. , Nature (1998) 282 902 -907

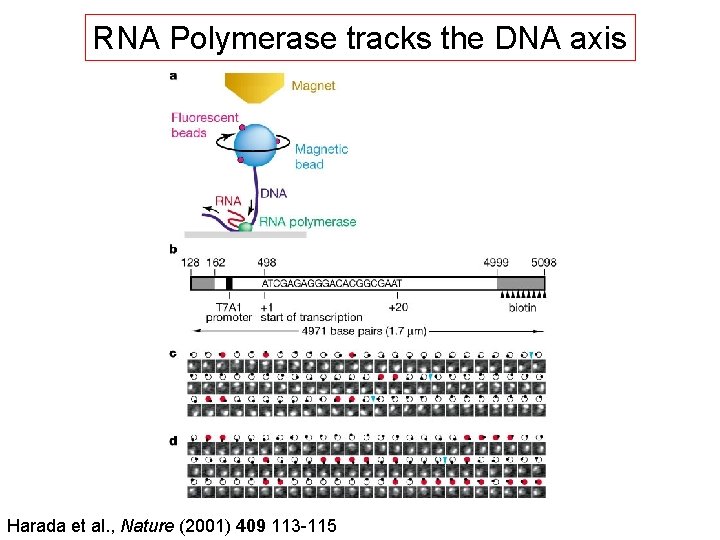
RNA Polymerase tracks the DNA axis Harada et al. , Nature (2001) 409 113 -115

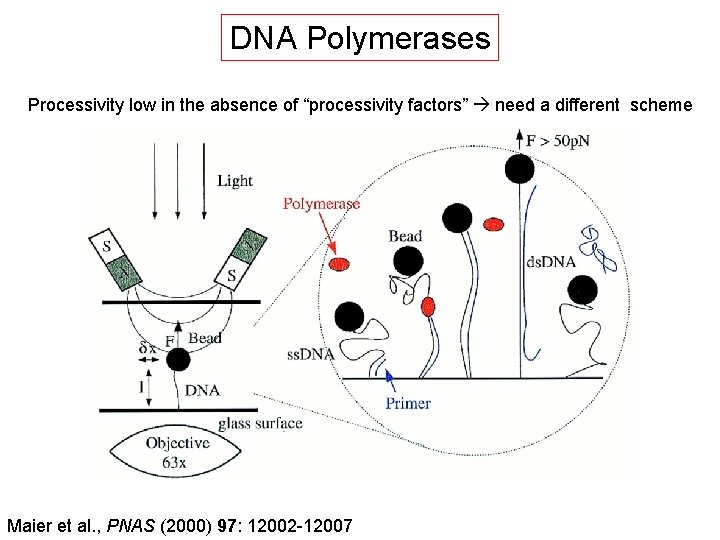
DNA Polymerases Processivity low in the absence of “processivity factors” need a different scheme Maier et al. , PNAS (2000) 97: 12002 -12007

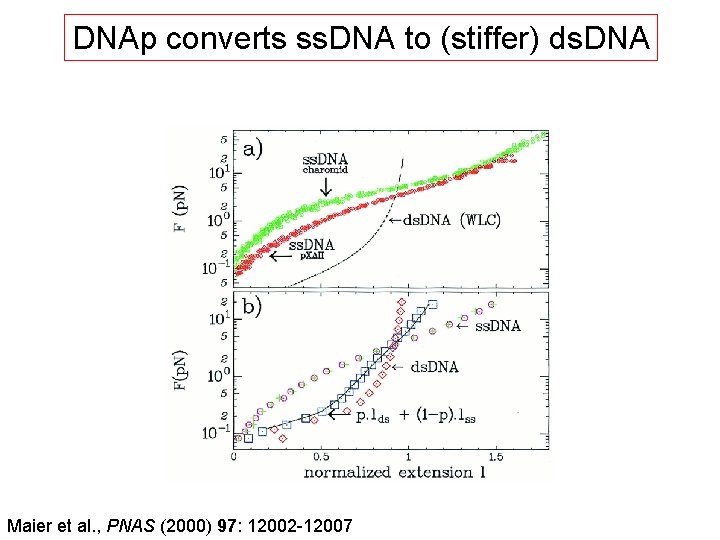
DNAp converts ss. DNA to (stiffer) ds. DNA Maier et al. , PNAS (2000) 97: 12002 -12007

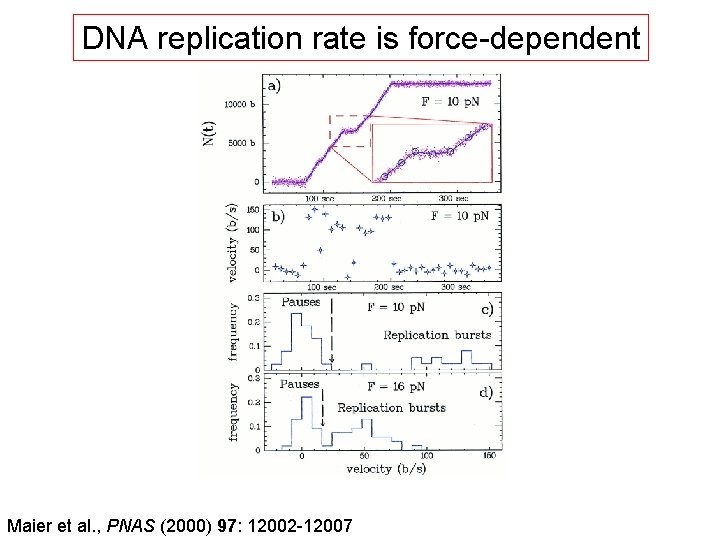
DNA replication rate is force-dependent Maier et al. , PNAS (2000) 97: 12002 -12007

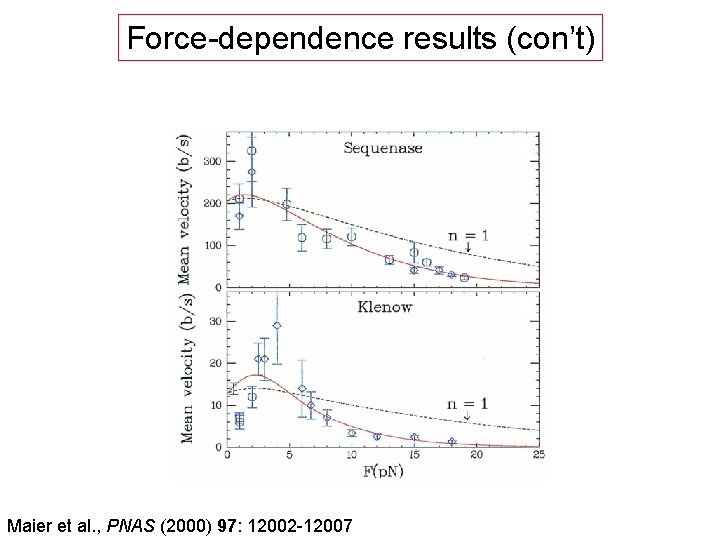
Force-dependence results (con’t) Maier et al. , PNAS (2000) 97: 12002 -12007

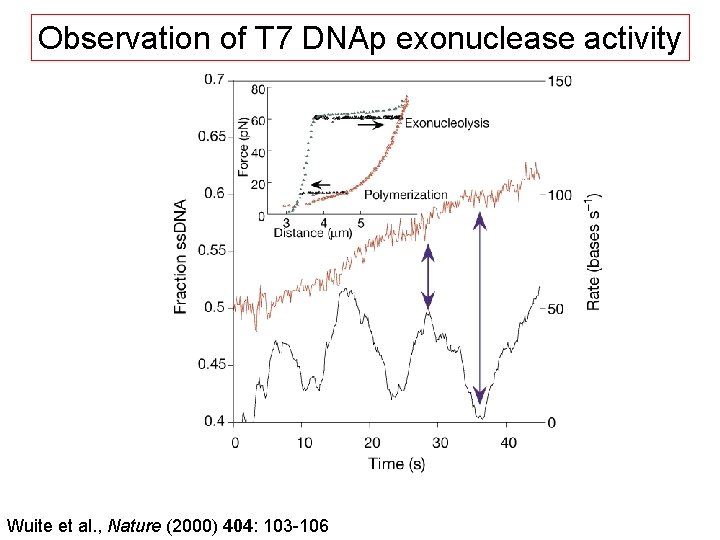
Observation of T 7 DNAp exonuclease activity Wuite et al. , Nature (2000) 404: 103 -106

Acknowledgements Rutgers Univ. A. Revyakin R. H. Ebright Research on transcription initiation funded by the Cold Spring Harbor Fellows program