Signalingmediated epigenetic silencing of tumor suppressor genes in





















- Slides: 48

Signaling-mediated epigenetic silencing of tumor suppressor genes in ovarian cancer Michael Chan, Ph. D. Associate Professor Department of Life Science National Chung Cheng University, Min Hsiung, Chia-Yi

Our Epigenome • Heritable changes that modulate gene expression without changes in DNA sequence – DNA methylation – Histone modifications – Nucleosome positioning – Chromatin remodeling – Non-coding RNA regulation

DNA Methylation • Occurs in CG dinucleotide (Cp. G) • Cp. G islands: CG rich region (500 bp-2000 bp) in the promoter region (of 50% human genes) • Repress transcription DNA methyltransferase (DNMT) S-adenosyl methionine (SAM) Demethylation (Tet, 5 hm. C) cytosine 5 -methylcytosine

Altered DNA-methylation patterns in tumorigenesis Esteller Nat Rev Genet 2007

Histone modification and histone codes Modification Histone Site Function Acetylation (Ac) H 3 Lysine 9 (K 9) activation Methylation (Me) H 3 Lysine 4 (K 4) activation Methylation (Me) H 3 Lysine 27 (K 27) repression Methylation (Me) H 3 Lysine 9 (K 9) repression

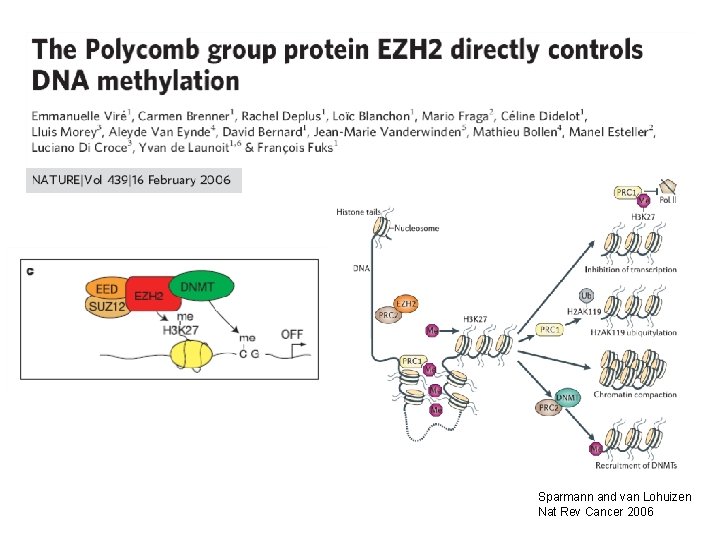
Sparmann and van Lohuizen Nat Rev Cancer 2006

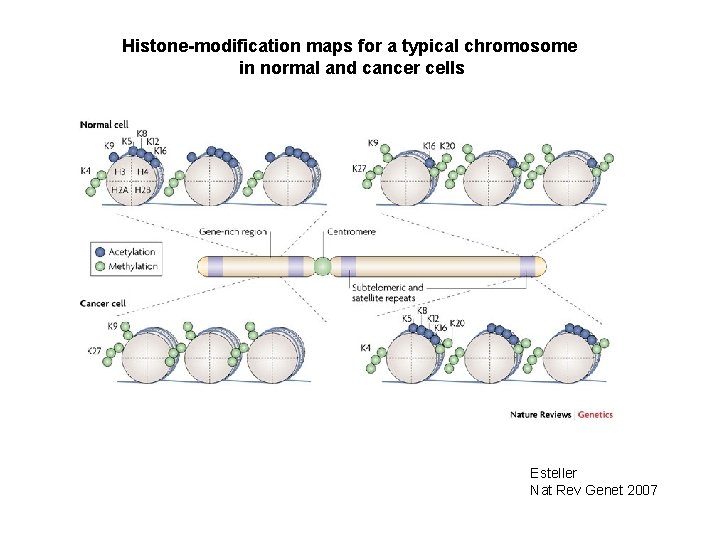
Histone-modification maps for a typical chromosome in normal and cancer cells Esteller Nat Rev Genet 2007

Hypothesis: Signaling-mediated epigenetic silencing

TGF-β signaling and ovarian cancer The Ovary

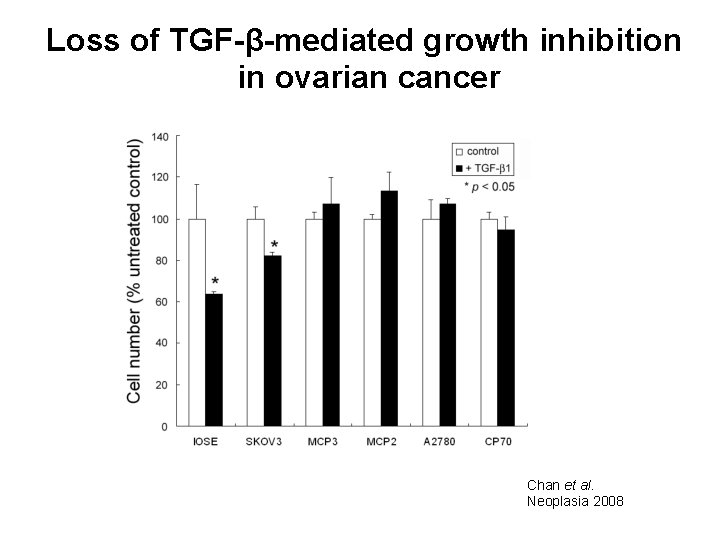
Loss of TGF-β-mediated growth inhibition in ovarian cancer Chan et al. Neoplasia 2008

Tumor suppressor Oncogenic Tumor progression Invasion metastasis TGF- Growth inhibition Apoptosis Genomic stability Master Yoda: “May the force be with you, but beware the dark side of the force”

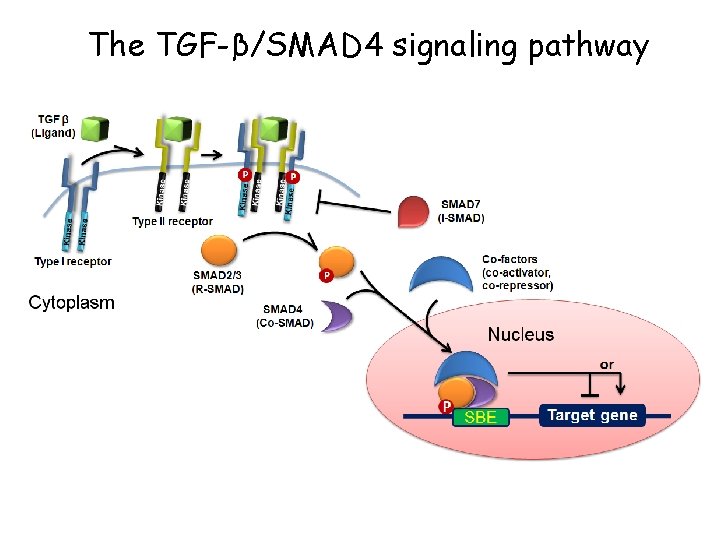
The TGF-β/SMAD 4 signaling pathway

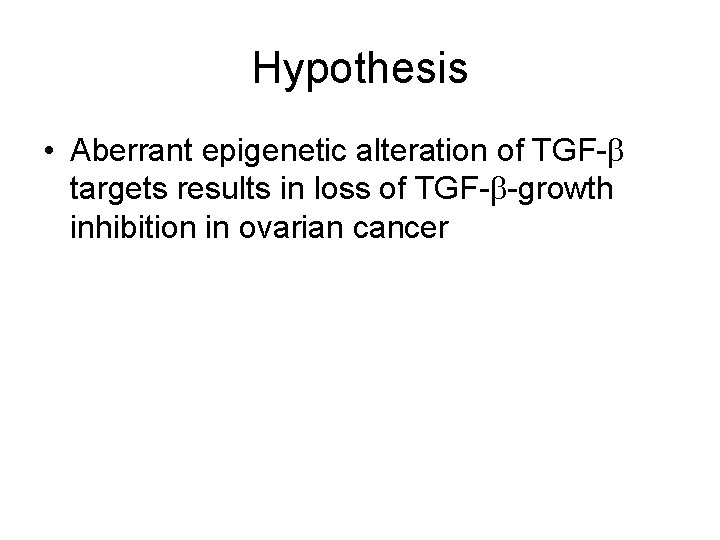
Hypothesis • Aberrant epigenetic alteration of TGF- targets results in loss of TGF- -growth inhibition in ovarian cancer

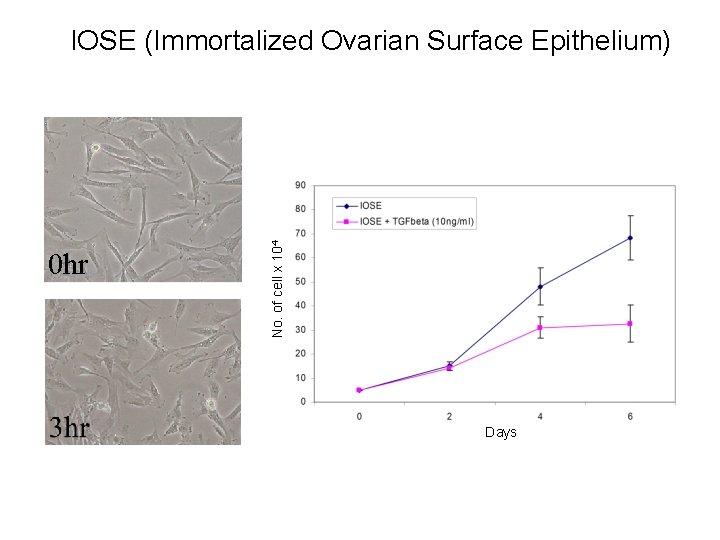
0 hr No. of cell x 104 IOSE (Immortalized Ovarian Surface Epithelium) Days

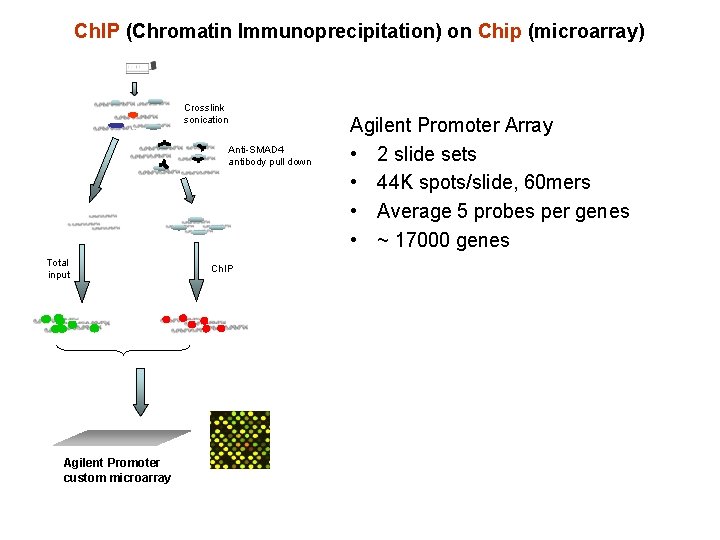
Ch. IP (Chromatin Immunoprecipitation) on Chip (microarray) Crosslink sonication Anti-SMAD 4 antibody pull down Total input Agilent Promoter custom microarray Ch. IP Agilent Promoter Array • 2 slide sets • 44 K spots/slide, 60 mers • Average 5 probes per genes • ~ 17000 genes

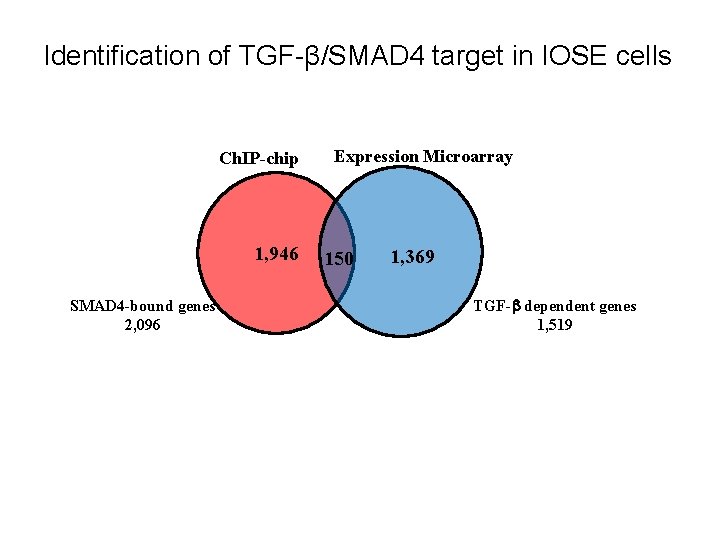
Identification of TGF-β/SMAD 4 target in IOSE cells Ch. IP-chip 1, 946 SMAD 4 -bound genes 2, 096 Expression Microarray 150 1, 369 TGF- dependent genes 1, 519

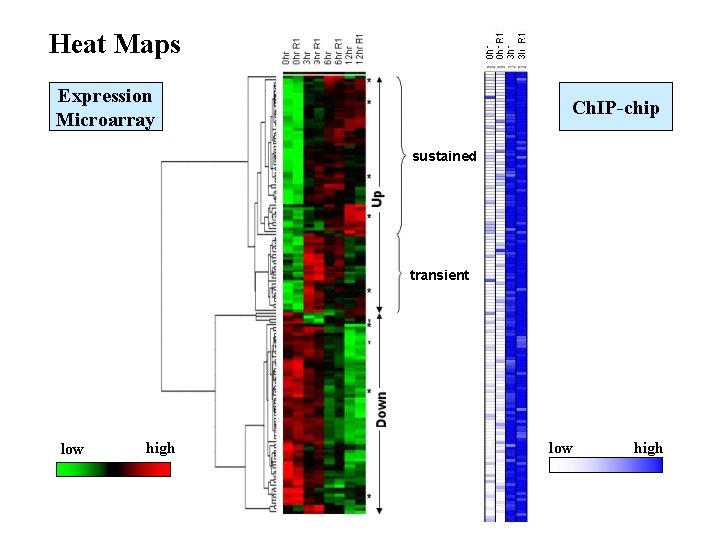
Heat Maps Expression Microarray Ch. IP-chip sustained transient low high

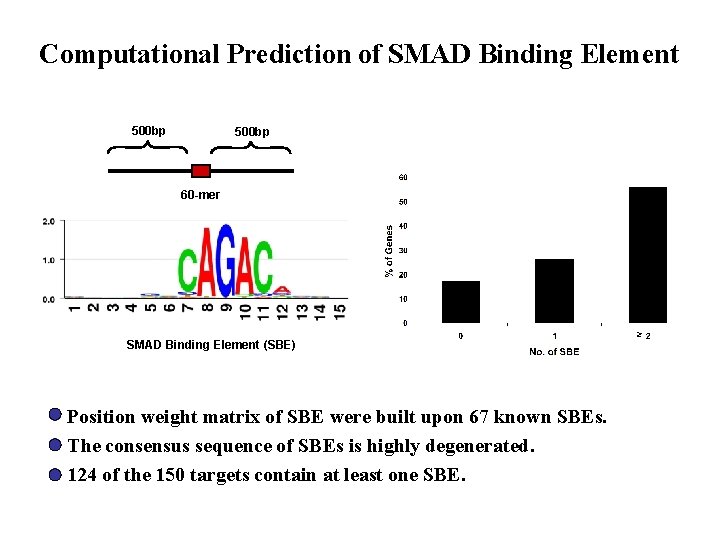
Computational Prediction of SMAD Binding Element 500 bp 60 -mer SMAD Binding Element (SBE) Position weight matrix of SBE were built upon 67 known SBEs. The consensus sequence of SBEs is highly degenerated. 124 of the 150 targets contain at least one SBE. ≥

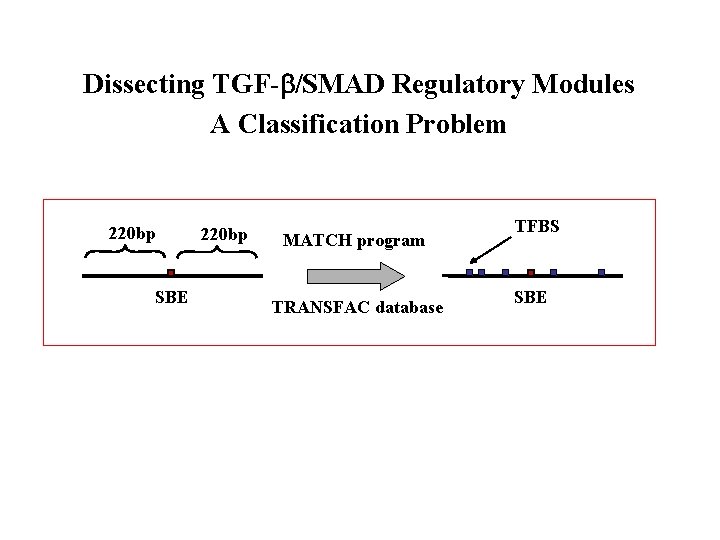
Dissecting TGF- /SMAD Regulatory Modules A Classification Problem 220 bp SBE 220 bp MATCH program TRANSFAC database TFBS SBE

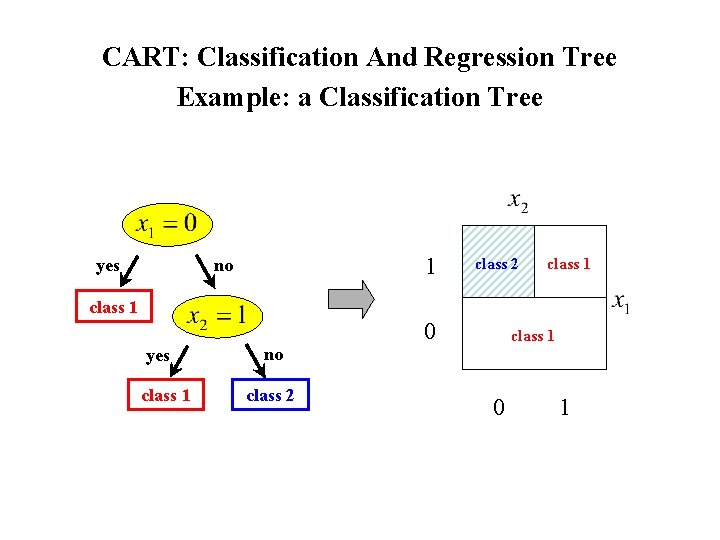
CART: Classification And Regression Tree Example: a Classification Tree 1 no yes class 2 class 1 0 yes class 1 no class 2 0 1

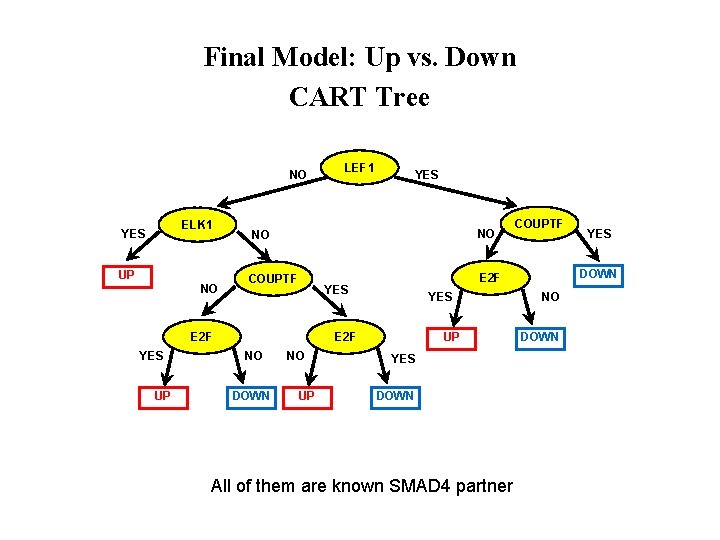
Final Model: Up vs. Down CART Tree NO ELK 1 YES UP NO UP YES NO NO COUPTF E 2 F YES LEF 1 YES UP E 2 F NO DOWN NO UP YES DOWN All of them are known SMAD 4 partner COUPTF YES DOWN NO DOWN

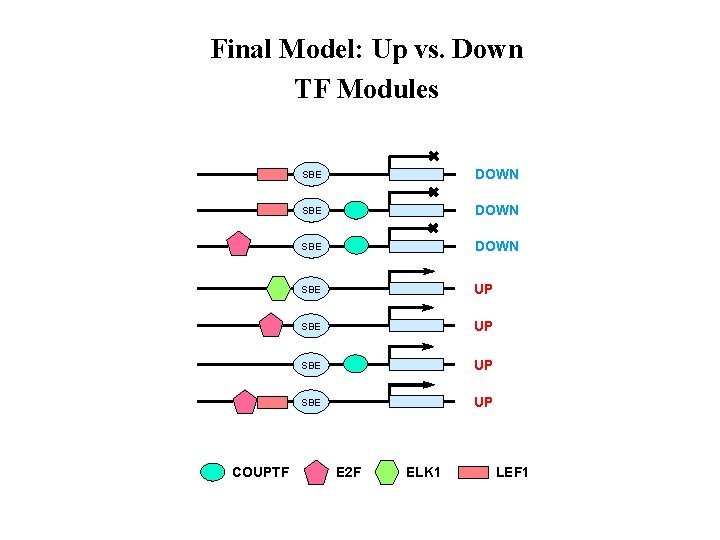
Final Model: Up vs. Down TF Modules COUPTF SBE DOWN SBE UP E 2 F ELK 1 LEF 1

Summary • Ch. IP-on-Chip couples with bioinformatics identifies 124 TGF- /SMAD 4 targets in IOSE cells • Bioinformatics predicts SMAD 4 regulatory modules in IOSE

Is SMAD 4 a master regulator of cancer methylome in ovarian cancer?


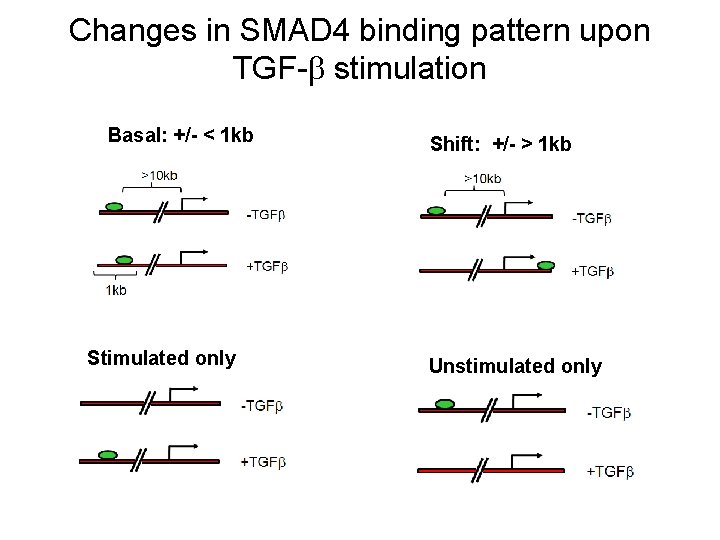
Changes in SMAD 4 binding pattern upon TGF- stimulation Basal: +/- < 1 kb Stimulated only Shift: +/- > 1 kb Unstimulated only

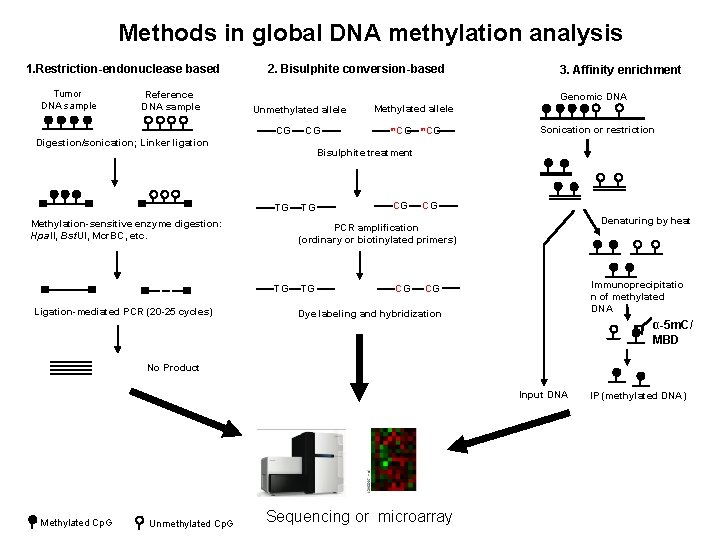
Methods in global DNA methylation analysis 1. Restriction-endonuclease based Tumor DNA sample Reference DNA sample 2. Bisulphite conversion-based Genomic DNA Unmethylated allele CG CG Digestion/sonication; Linker ligation Methylated allele m. CG Sonication or restriction Bisulphite treatment TG Methylation-sensitive enzyme digestion: Hpa. II, Bst. UI, Mcr. BC, etc. TG CG CG Denaturing by heat PCR amplification (ordinary or biotinylated primers) TG Ligation-mediated PCR (20 -25 cycles) 3. Affinity enrichment TG CG Immunoprecipitatio n of methylated DNA CG Dye labeling and hybridization α-5 m. C/ MBD No Product Input DNA Methylated Cp. G Unmethylated Cp. G Sequencing or microarray IP (methylated DNA)


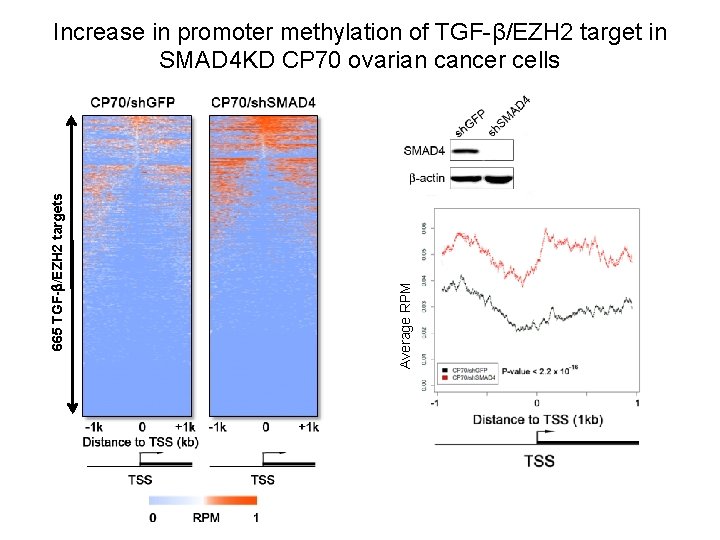
Methylation changes of SMAD 4 targets in SMAD 4 KD cells by Methyl. Cap-Seq Genomic DNA Sonication or restriction Immunoprecipitation of methylated DNA MBD RPM: read per millon (methylation level) Input DNA IP (methylated DNA)

Average RPM 665 TGF- /EZH 2 targets Increase in promoter methylation of TGF-β/EZH 2 target in SMAD 4 KD CP 70 ovarian cancer cells

Mathematical model of bimodal epigenetic control of mi. R-193 a in ovarian cancer

Ovarian cancer Lancet 2007; 369: 1703 -10 • Epidemiological studies found that women with hormone (estrogen) replacement therapy are associated with the risk of ovarian cancer. • However, anti-estrogen therapy is only partially effective in ovarian cancer. (Smyth et al. , Clin Cancer Res 2007)

CD 117 (c-kit) as a CIC marker in ovarian cancer

How’s CD 117 (c-kit) regulated epigenetically in ovarian cancer

mi. R-193 a targets c-kit 3’UTR mi. R-193 a 3’ ugac. CCUGAAACAUCCGGUCAa 5’ mi. R-193 a AAA… c-KIT m. RNA (3’UTR 2158 bp) = mi. R-193 a binding site 5’ uccu. GGACACCG--GGCCAGUa 3’ c-KIT mi. R 193 a MRE in c-KIT is 1075 bp from 3’UTR start

Chen et al, Nature Rev Cancer 2009

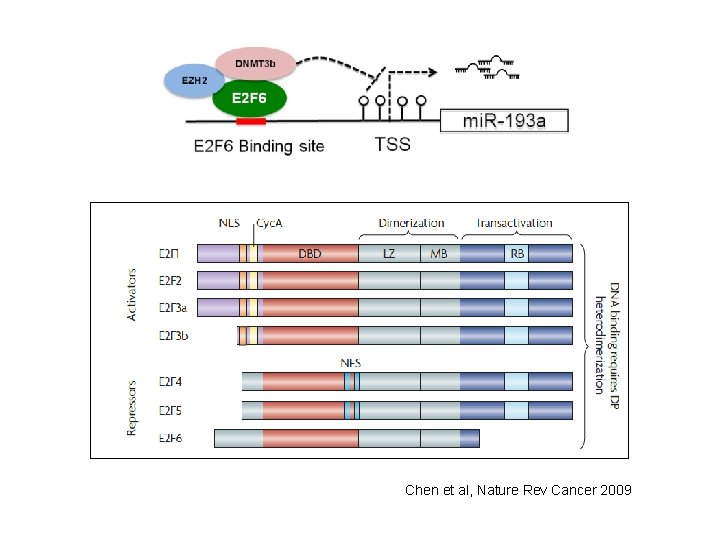
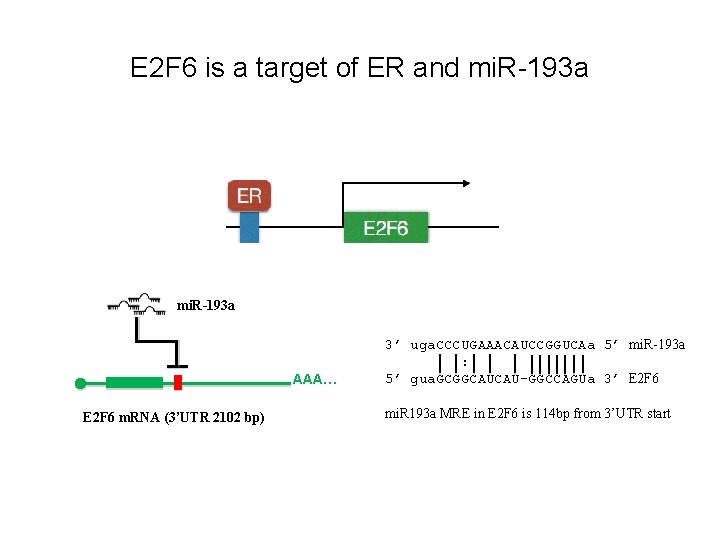
E 2 F 6 is a target of ER and mi. R-193 a AAA… E 2 F 6 m. RNA (3’UTR 2102 bp) 3’ uga. CCCUGAAACAUCCGGUCAa 5’ mi. R-193 a : 5’ gua. GCGGCAUCAU-GGCCAGUa 3’ E 2 F 6 mi. R 193 a MRE in E 2 F 6 is 114 bp from 3’UTR start

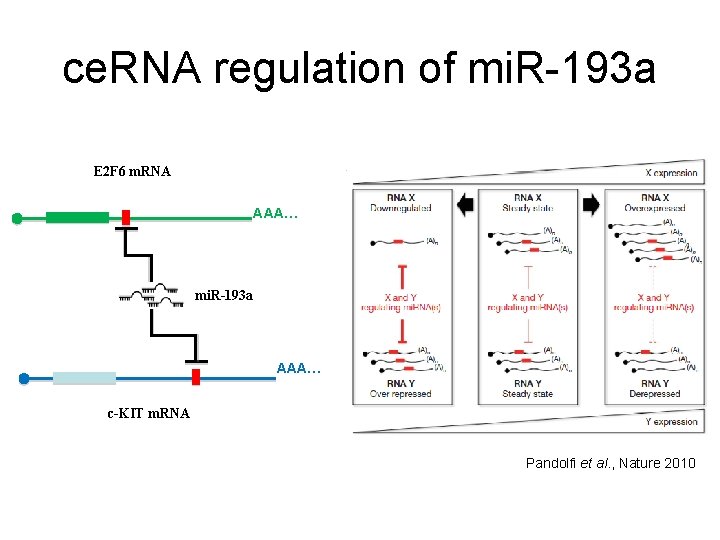
ce. RNA regulation of mi. R-193 a E 2 F 6 m. RNA AAA… mi. R-193 a AAA… c-KIT m. RNA Pandolfi et al. , Nature 2010

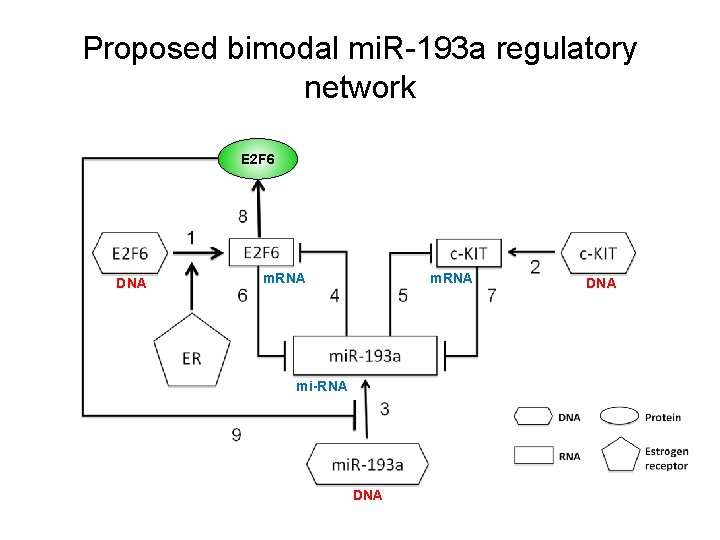
Proposed bimodal mi. R-193 a regulatory network E 2 F 6 DNA m. RNA mi-RNA DNA

Mathematical modeling of mi. R-193 a regulatory network k 5: transcription rate of E 2 F 6 Rc: amount of c-kit m. RNA K 4: strength of transcriptional inhibition (inhibition rate) by E 2 F 6

amount of c-kit m. RNA Steady-state bifurcation diagram inhibition rate of E 2 F 6 transcription rate of E 2 F 6

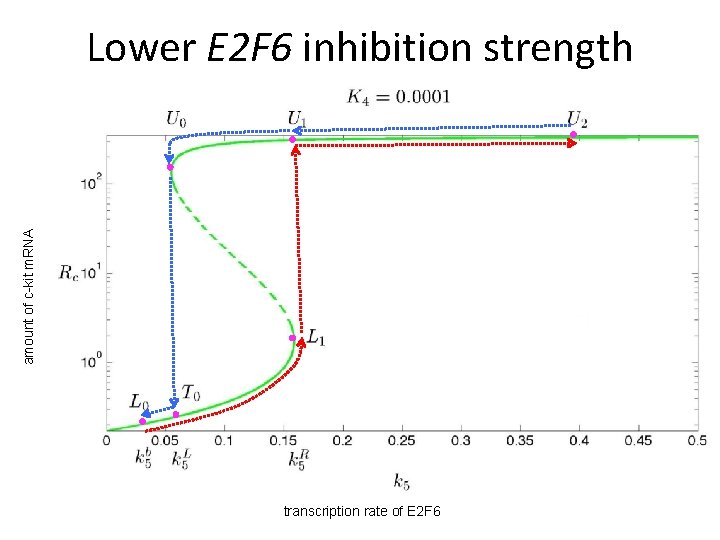
amount of c-kit m. RNA Lower E 2 F 6 inhibition strength transcription rate of E 2 F 6

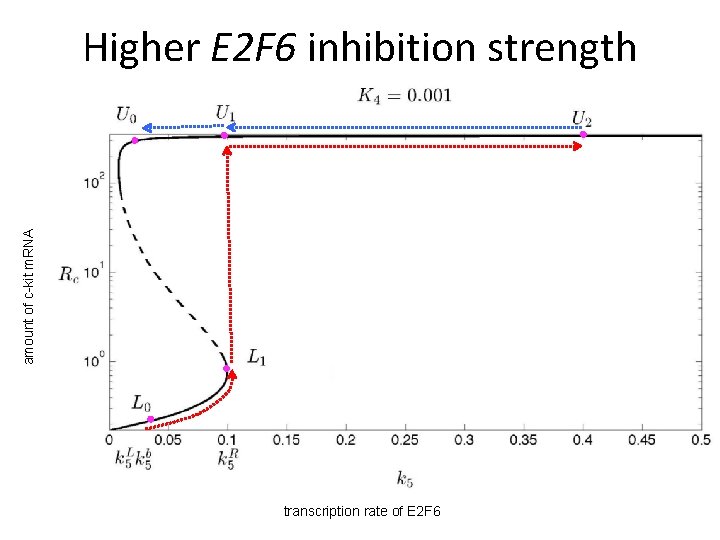
amount of c-kit m. RNA Higher E 2 F 6 inhibition strength transcription rate of E 2 F 6

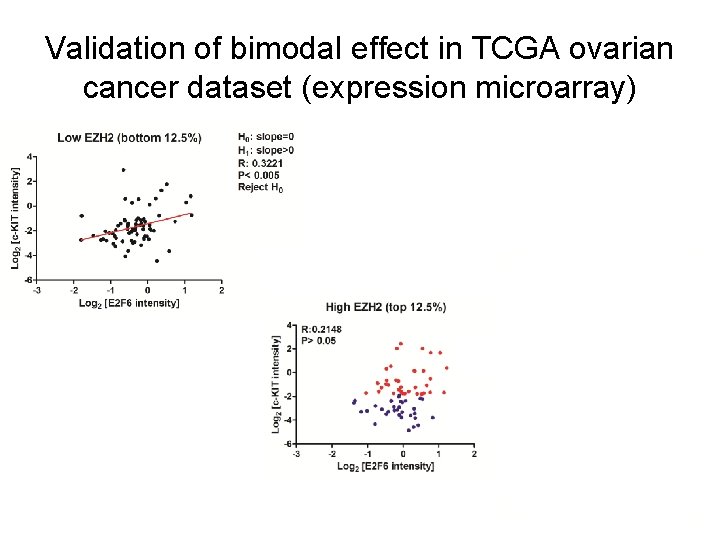
Validation of bimodal effect in TCGA ovarian cancer dataset (expression microarray)

Our model Low K 4 due to low EZH 2 level High K 4 due to high EZH 2 level


Acknowledgements National Chung Cheng University Life Science Jian-Liang Chou Frank Cheng Alex Chang Li-Han Zeng Lin-Yu Chen Mathematics Je-Chiang Tsai Tzy-Wei Huang Computer Science Yau-Ting Huang Taipei Medical University Hung-Cheng Lai UTHSC, San Antonio, Texas Tim H. -M. Huang Victor X. Jin The Ohio State University Shili Lin Pearlly S. Yan Jiayuh Lin Indiana University Kenneth P. Nephew Philippine Genome Center Baltazar D. Aguda Polish Academy of Science Marek Kochańczyk

~ THE END ~ Thank you for your attention!