Short Read Sequencing Analysis Workshop Day 1 Considerations

- Slides: 26

Short Read Sequencing Analysis Workshop Day 1 Considerations for Sequencing

Different types of sequencing libraries • • Whole genome sequencing RNA Sequencing/GRO-Seq Ch. IP-seq DNAse 1, ATAC-seq Exome sequencing Methyl-Seq Metagenomic/Amplicon (low diversity)

Platform Comparsion

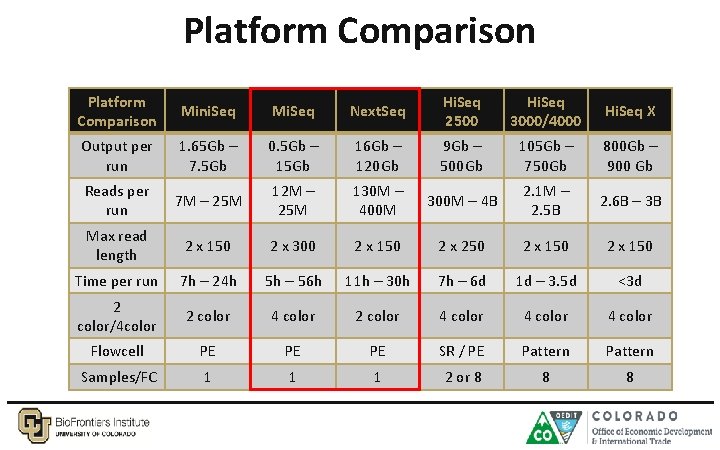
Platform Comparison Mini. Seq Mi. Seq Next. Seq Hi. Seq 2500 Hi. Seq 3000/4000 Hi. Seq X Output per run 1. 65 Gb – 7. 5 Gb 0. 5 Gb – 15 Gb 16 Gb – 120 Gb 9 Gb – 500 Gb 105 Gb – 750 Gb 800 Gb – 900 Gb Reads per run 7 M – 25 M 12 M – 25 M 130 M – 400 M 300 M – 4 B 2. 1 M – 2. 5 B 2. 6 B – 3 B Max read length 2 x 150 2 x 300 2 x 150 2 x 250 2 x 150 Time per run 7 h – 24 h 5 h – 56 h 11 h – 30 h 7 h – 6 d 1 d – 3. 5 d <3 d 2 color/4 color 2 color 4 color Flowcell PE PE PE SR / PE Pattern Samples/FC 1 1 1 2 or 8 8 8

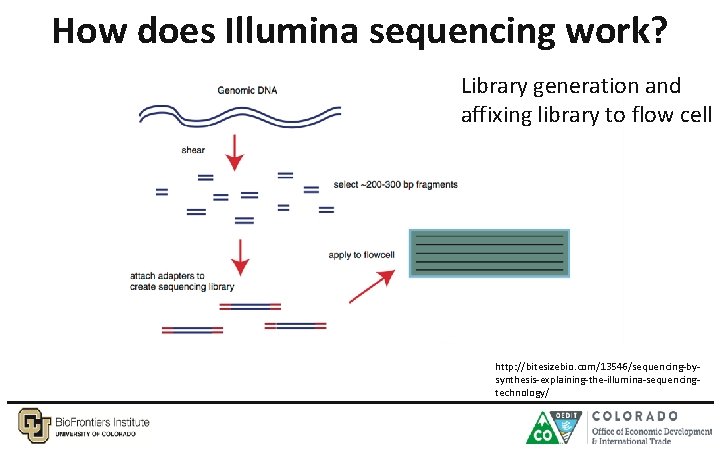
How does Illumina sequencing work? Library generation and affixing library to flow cell http: //bitesizebio. com/13546/sequencing-bysynthesis-explaining-the-illumina-sequencingtechnology/

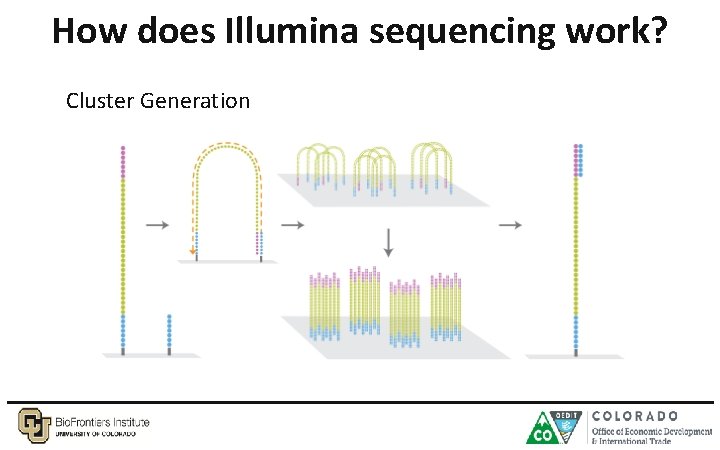
How does Illumina sequencing work? Cluster Generation

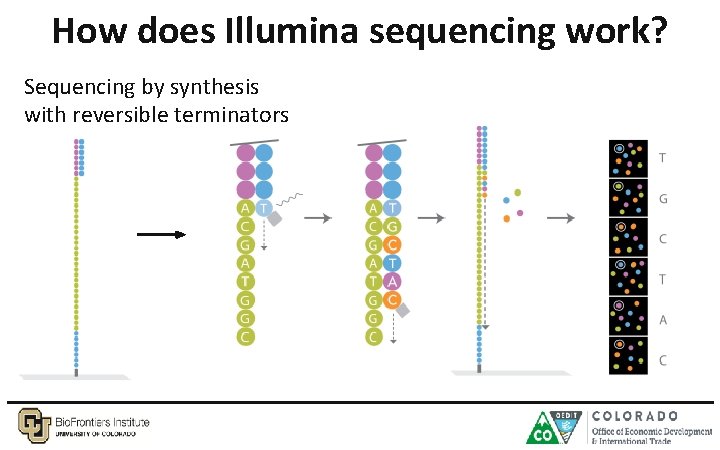
How does Illumina sequencing work? Sequencing by synthesis with reversible terminators

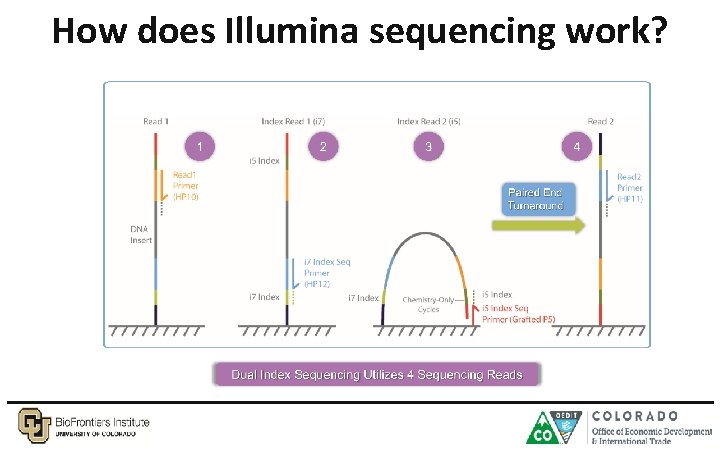
How does Illumina sequencing work?

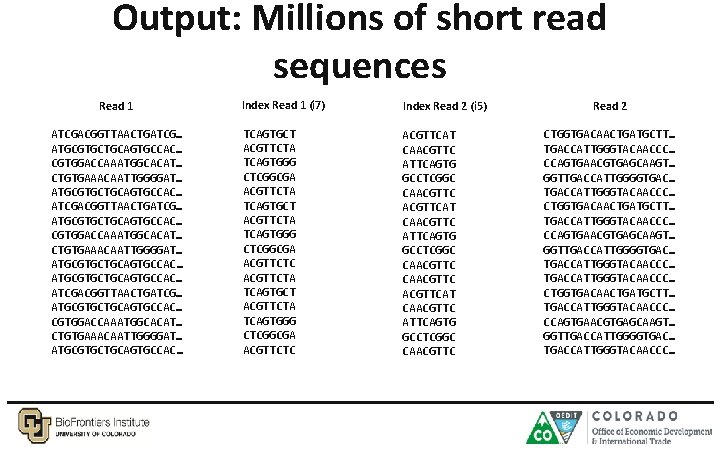
Output: Millions of short read sequences Read 1 ATCGACGGTTAACTGATCG… ATGCGTGCTGCAGTGCCAC… CGTGGACCAAATGGCACAT… CTGTGAAACAATTGGGGAT… ATGCGTGCTGCAGTGCCAC… ATCGACGGTTAACTGATCG… ATGCGTGCTGCAGTGCCAC… CGTGGACCAAATGGCACAT… CTGTGAAACAATTGGGGAT… ATGCGTGCTGCAGTGCCAC… Index Read 1 (i 7) Index Read 2 (i 5) TCAGTGCT ACGTTCTA TCAGTGGG CTCGGCGA ACGTTCTA TCAGTGCT ACGTTCTA TCAGTGGG CTCGGCGA ACGTTCTC ACGTTCAT CAACGTTC ATTCAGTG GCCTCGGC CAACGTTC ACGTTCAT CAACGTTC ATTCAGTG GCCTCGGC CAACGTTC Read 2 CTGGTGACAACTGATGCTT… TGACCATTGGGTACAACCC… CCAGTGAACGTGAGCAAGT… GGTTGACCATTGGGGTGAC… TGACCATTGGGTACAACCC… CTGGTGACAACTGATGCTT… TGACCATTGGGTACAACCC… CCAGTGAACGTGAGCAAGT… GGTTGACCATTGGGGTGAC… TGACCATTGGGTACAACCC…

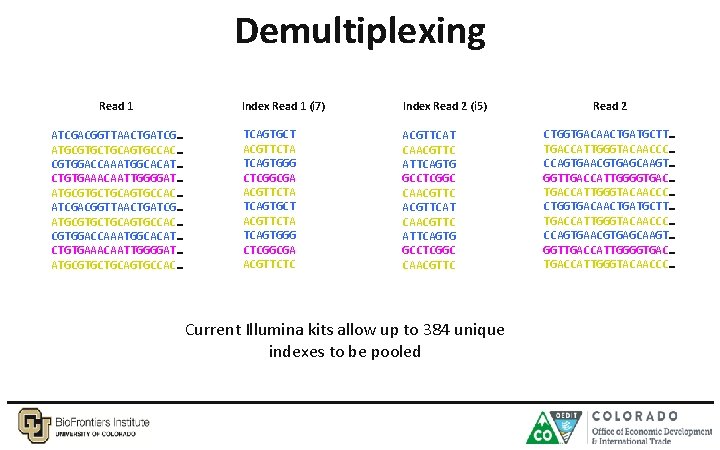
Demultiplexing Read 1 ATCGACGGTTAACTGATCG… ATGCGTGCTGCAGTGCCAC… CGTGGACCAAATGGCACAT… CTGTGAAACAATTGGGGAT… ATGCGTGCTGCAGTGCCAC… Index Read 1 (i 7) Index Read 2 (i 5) TCAGTGCT ACGTTCTA TCAGTGGG CTCGGCGA ACGTTCTA TCAGTGCT ACGTTCTA TCAGTGGG CTCGGCGA ACGTTCTC ACGTTCAT CAACGTTC ATTCAGTG GCCTCGGC CAACGTTC Current Illumina kits allow up to 384 unique indexes to be pooled Read 2 CTGGTGACAACTGATGCTT… TGACCATTGGGTACAACCC… CCAGTGAACGTGAGCAAGT… GGTTGACCATTGGGGTGAC… TGACCATTGGGTACAACCC…

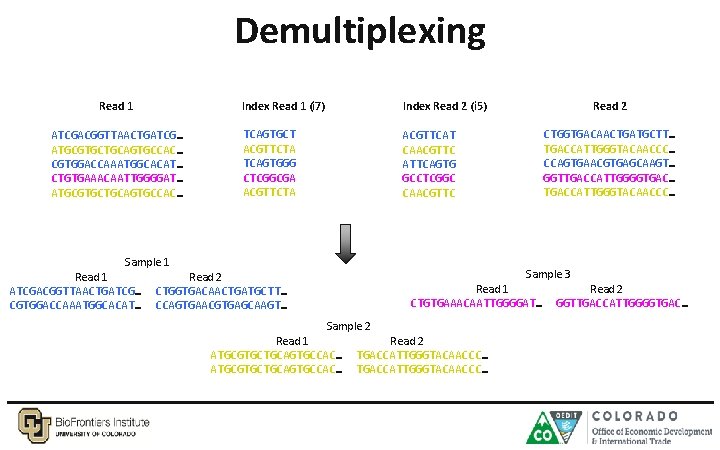
Demultiplexing Read 1 ATCGACGGTTAACTGATCG… ATGCGTGCTGCAGTGCCAC… CGTGGACCAAATGGCACAT… CTGTGAAACAATTGGGGAT… ATGCGTGCTGCAGTGCCAC… Index Read 1 (i 7) Index Read 2 (i 5) TCAGTGCT ACGTTCTA TCAGTGGG CTCGGCGA ACGTTCTA ACGTTCAT CAACGTTC ATTCAGTG GCCTCGGC CAACGTTC Sample 1 Read 1 ATCGACGGTTAACTGATCG… CGTGGACCAAATGGCACAT… Read 2 CTGGTGACAACTGATGCTT… TGACCATTGGGTACAACCC… CCAGTGAACGTGAGCAAGT… GGTTGACCATTGGGGTGAC… TGACCATTGGGTACAACCC… Sample 3 Read 2 CTGGTGACAACTGATGCTT… CCAGTGAACGTGAGCAAGT… Read 1 CTGTGAAACAATTGGGGAT… Sample 2 Read 1 ATGCGTGCTGCAGTGCCAC… Read 2 TGACCATTGGGTACAACCC… Read 2 GGTTGACCATTGGGGTGAC…

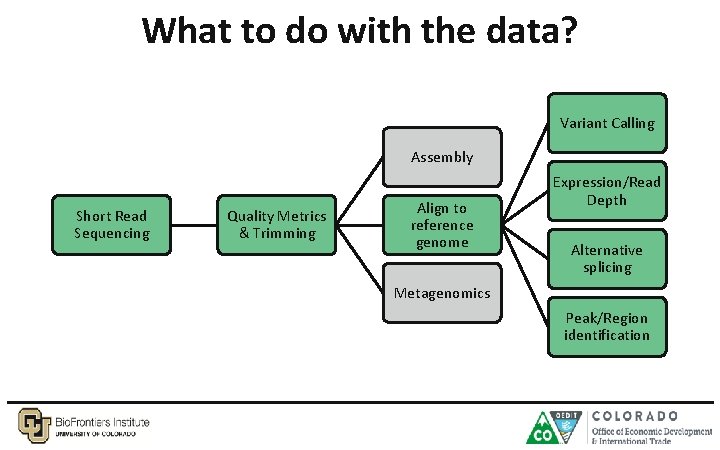
What to do with the data? Variant Calling Assembly Short Read Sequencing Quality Metrics & Trimming Align to reference genome Expression/Read Depth Alternative splicing Metagenomics Peak/Region identification

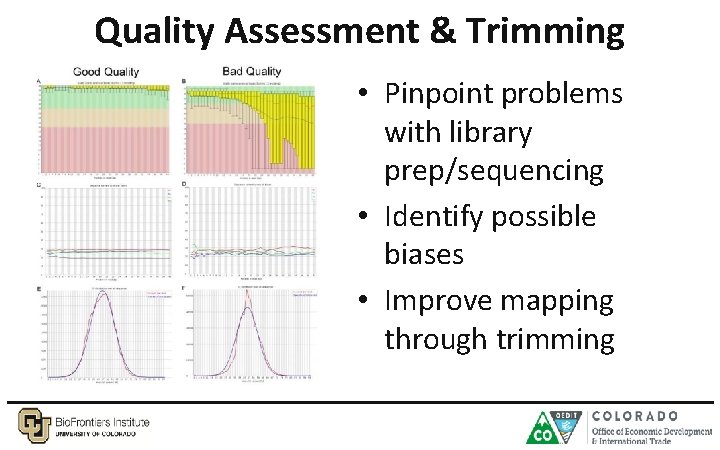
Quality Assessment & Trimming • Pinpoint problems with library prep/sequencing • Identify possible biases • Improve mapping through trimming

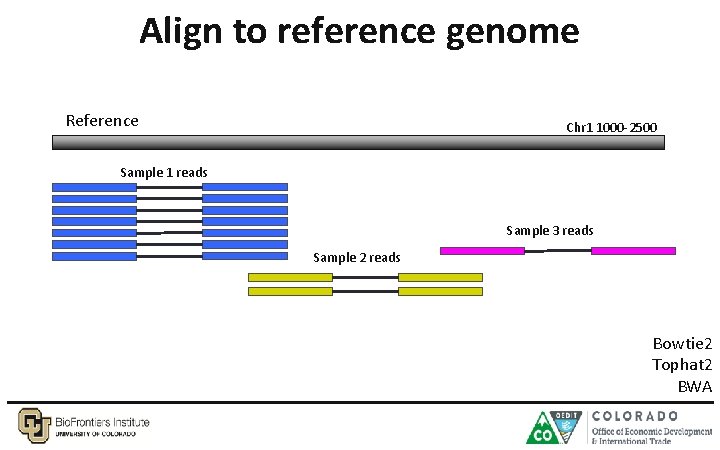
Align to reference genome Reference Chr 1 1000 -2500 Sample 1 reads Sample 3 reads Sample 2 reads Bowtie 2 Tophat 2 BWA

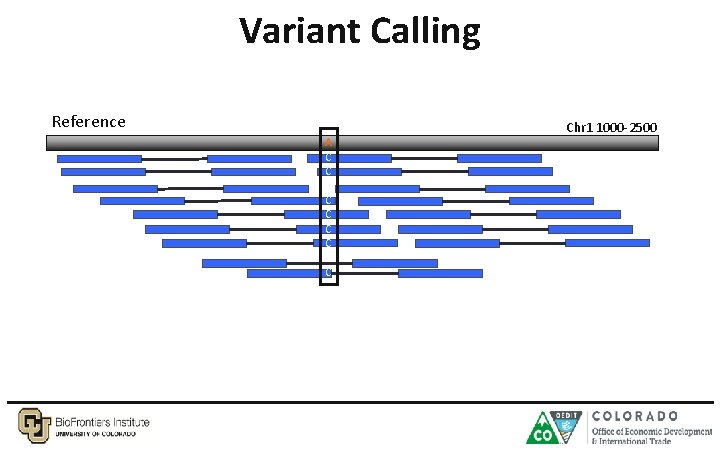
Variant Calling Reference A C C C Chr 1 1000 -2500

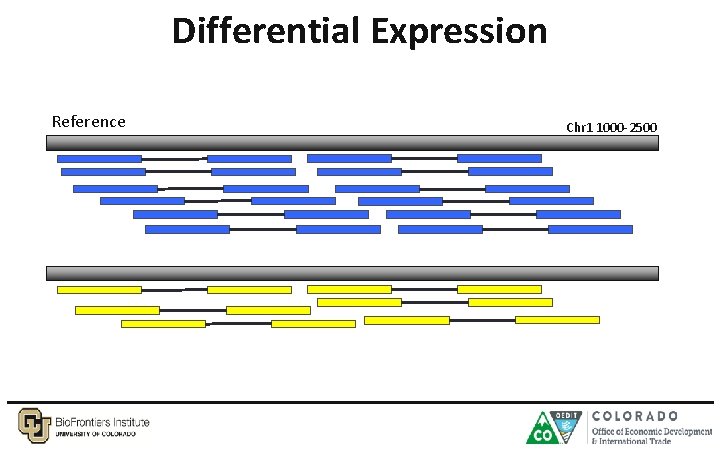
Differential Expression Reference Chr 1 1000 -2500

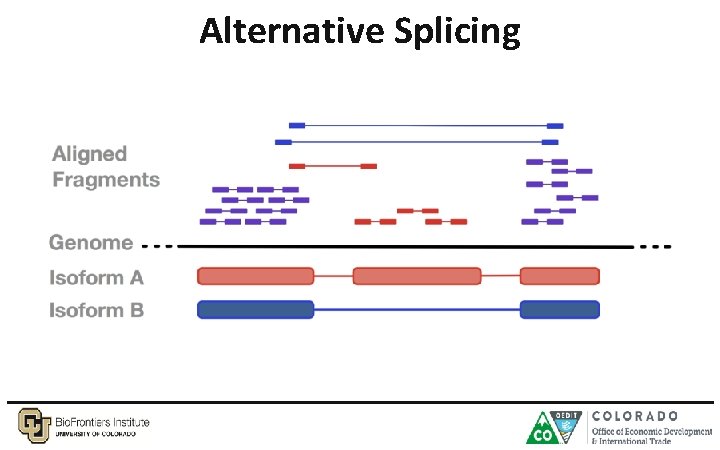
Alternative Splicing

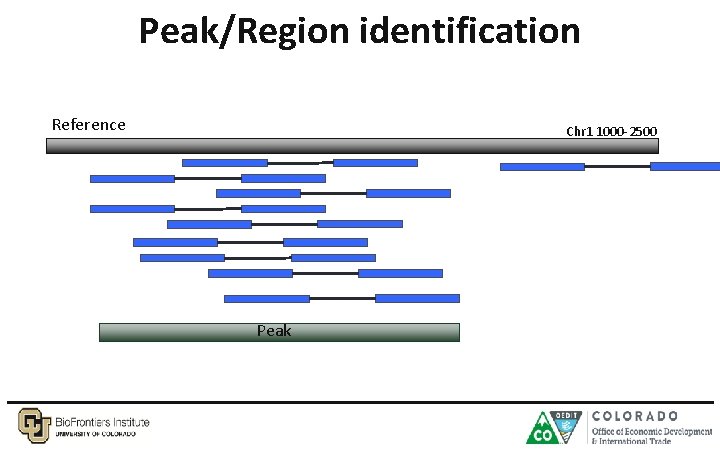
Peak/Region identification Reference Chr 1 1000 -2500 Peak

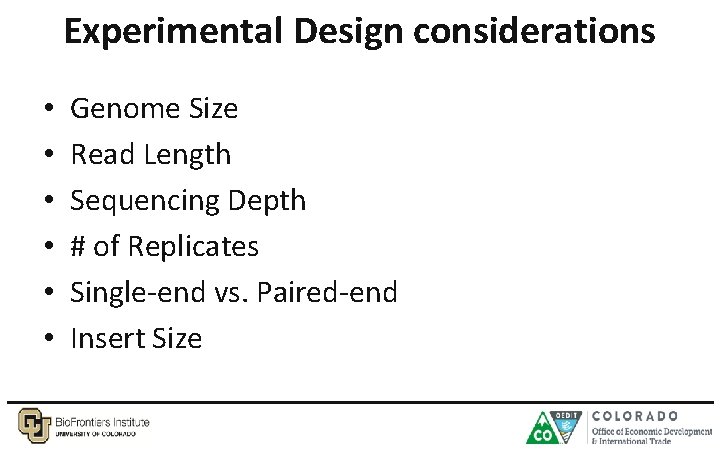
Experimental Design considerations • • • Genome Size Read Length Sequencing Depth # of Replicates Single-end vs. Paired-end Insert Size

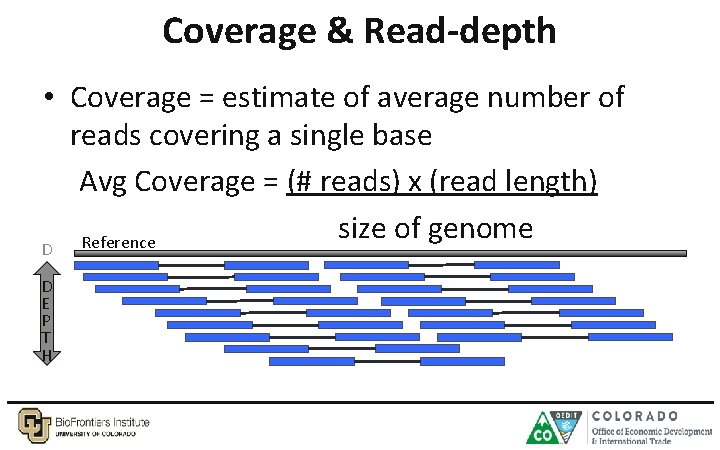
Coverage & Read-depth • Coverage = estimate of average number of reads covering a single base Avg Coverage = (# reads) x (read length) size of genome Reference D p E Pt Th H

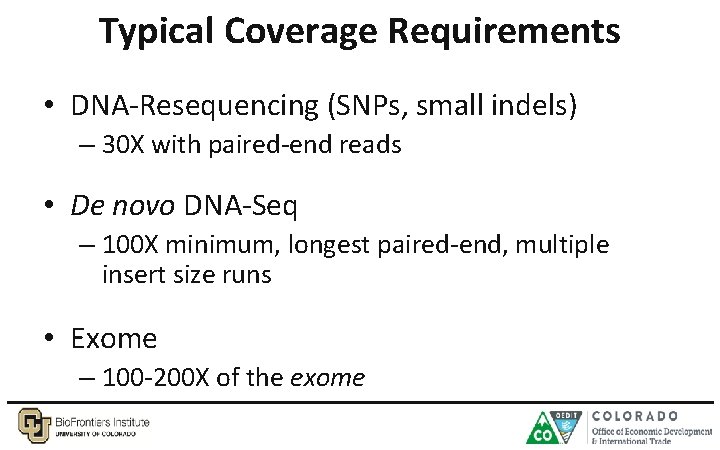
Typical Coverage Requirements • DNA-Resequencing (SNPs, small indels) – 30 X with paired-end reads • De novo DNA-Seq – 100 X minimum, longest paired-end, multiple insert size runs • Exome – 100 -200 X of the exome

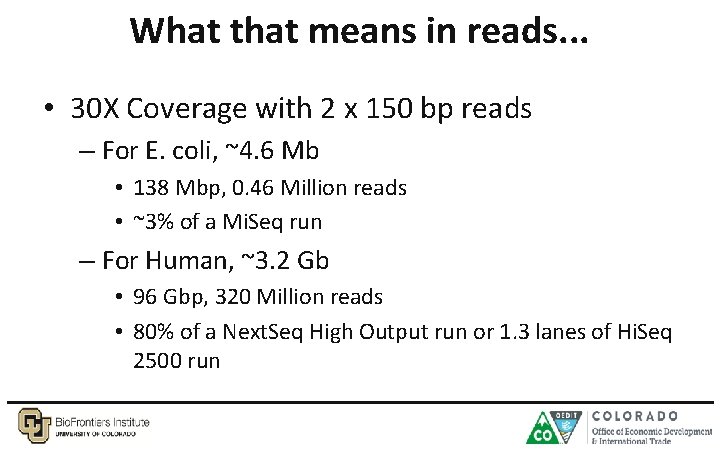
What that means in reads. . . • 30 X Coverage with 2 x 150 bp reads – For E. coli, ~4. 6 Mb • 138 Mbp, 0. 46 Million reads • ~3% of a Mi. Seq run – For Human, ~3. 2 Gb • 96 Gbp, 320 Million reads • 80% of a Next. Seq High Output run or 1. 3 lanes of Hi. Seq 2500 run

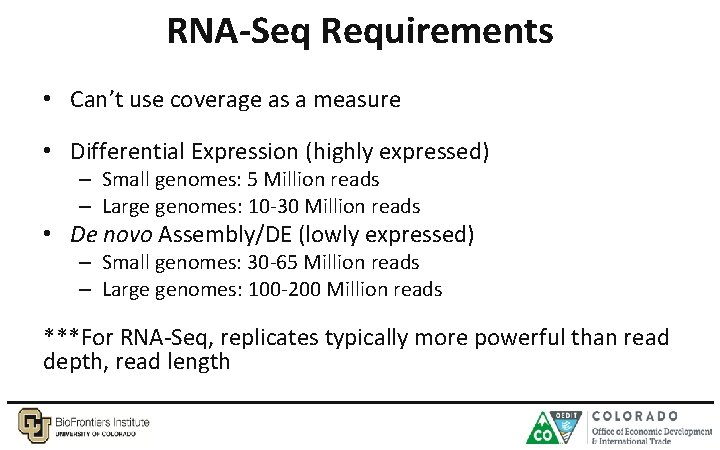
RNA-Seq Requirements • Can’t use coverage as a measure • Differential Expression (highly expressed) – Small genomes: 5 Million reads – Large genomes: 10 -30 Million reads • De novo Assembly/DE (lowly expressed) – Small genomes: 30 -65 Million reads – Large genomes: 100 -200 Million reads ***For RNA-Seq, replicates typically more powerful than read depth, read length

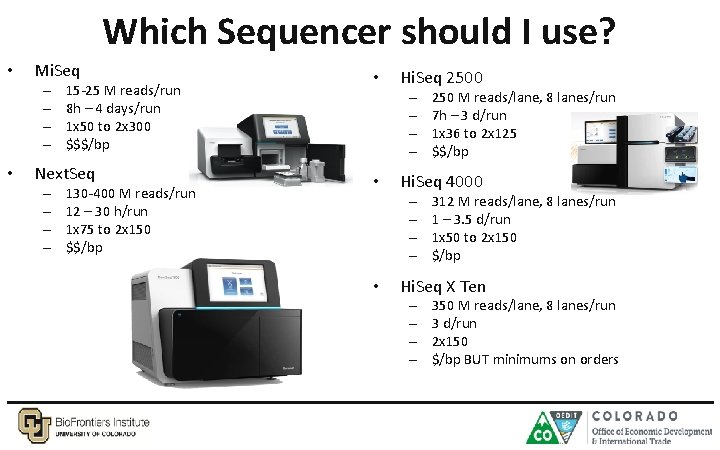
Which Sequencer should I use? • Mi. Seq – – • 15 -25 M reads/run 8 h – 4 days/run 1 x 50 to 2 x 300 $$$/bp Next. Seq – – 130 -400 M reads/run 12 – 30 h/run 1 x 75 to 2 x 150 $$/bp • Hi. Seq 2500 – – • Hi. Seq 4000 – – • 250 M reads/lane, 8 lanes/run 7 h – 3 d/run 1 x 36 to 2 x 125 $$/bp 312 M reads/lane, 8 lanes/run 1 – 3. 5 d/run 1 x 50 to 2 x 150 $/bp Hi. Seq X Ten – – 350 M reads/lane, 8 lanes/run 3 d/run 2 x 150 $/bp BUT minimums on orders

Other considerations • • • Base diversity (at each position) Custom versus kitted libraries – kit biases PCR/PCR-free libraries How unique is the run-type you want Queue times/Data delivery times Many more. .

Questions?