Setting up visualization Make output folder for visualization



























- Slides: 30

Setting up visualization

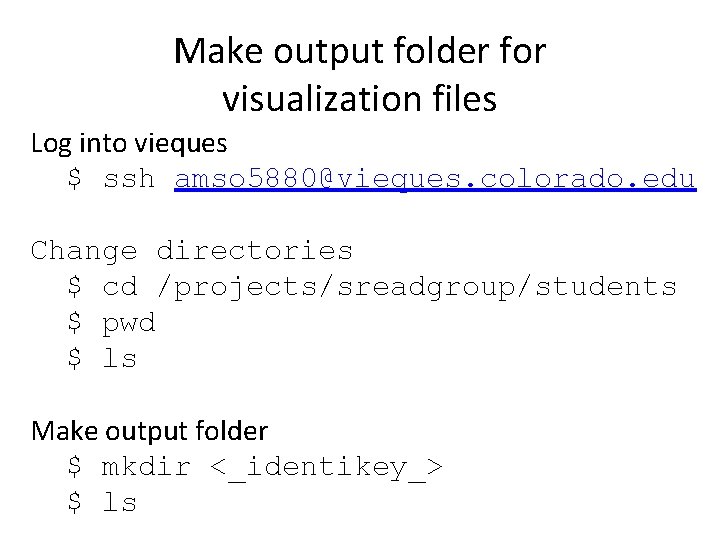
Make output folder for visualization files Log into vieques $ ssh amso 5880@vieques. colorado. edu Change directories $ cd /projects/sreadgroup/students $ pwd $ ls Make output folder $ mkdir <_identikey_> $ ls

Follow directions for setting up your vnc session • Check email from Jamie Kershner for port and link to setup vnc session http: //biof-edu. colorado. edu/videos/dowellshort-read-class/visualization-information

What do I do when I get data back from the sequencer?


Prep your home directory $ cd /Users/identikey/ $ mkdir Day 4 $ cd Day 4 $ mkdir fastq $ mkdir PBS/eofiles/

Copy today’s files $ scp /projects/sreadgroup/Day 4/fastq/* /Users/identikey/Day 4/fastq $ cp /projects/sreadgroup/Day 4/PBS/* /Users/identikey/Day 4/PBS

Unzip your files $ cd /Users/identikey/Day 4/fastq $ ls $ gunzip * $ ls $ less Example_A_01. fastq

What is a Fastq file? Read Identifier Sequence Quality

For the remainder of the Fast. QC examples, the class will be split into 4 groups • • • Group A – Work with Example_A files Group B – Work with Example_B files Group C – Work with Example_C files Group D – Work with Example_D files DO NOT DELETE the other files from your folder, they will be used in your homework

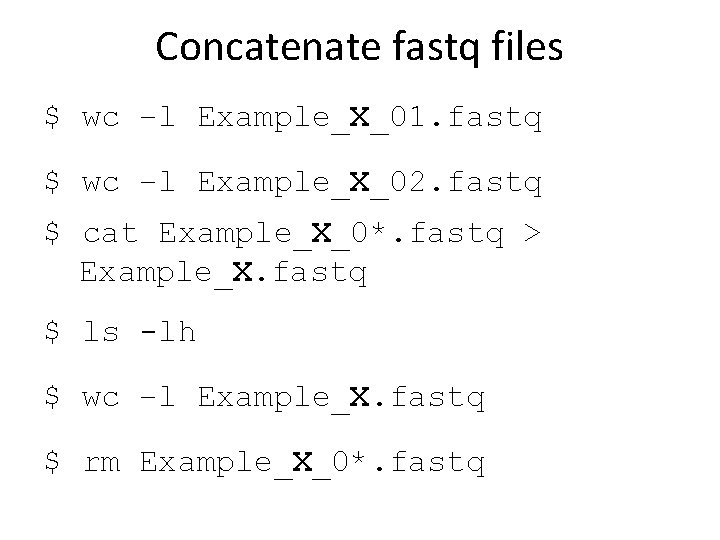
Concatenate fastq files $ wc –l Example_X_01. fastq $ wc –l Example_X_02. fastq $ cat Example_X_0*. fastq > Example_X. fastq $ ls -lh $ wc –l Example_X. fastq $ rm Example_X_0*. fastq

Evaluating Sequencing and Library Quality

Where to find Fast. QC options $ /opt/fast. QC/fastqc --help -OR$ module load fastqc_0. 11. 2 $ fastqc --help Don’t know how to use a program? Look at the help page for options!

Running Fast. QC $ cd /Users/identikey/Day 4/PBS/ $ ls $ nano run_fastqc. pbs

Replace anything with "<___>" with your desired value & "amso 5880" with your identikey • Specify Job name • Update resources if necessary • Specify Queue • Update eo file path #PBS –e /Users/identikey/Day 4/PBS/eofiles/ #PBS –o /Users/identikey/Day 4/PBS/eofiles/ • Update email • Specify output dir -o /Users/identikey/Day 4/QC/ • Specify input file /Users/identikey/Day 4/Example_X. fastq

Submit Fast. QC job $ $ module list module load fastqc_0. 11. 2 qsub run_fastqc. pbs qstat –u identikey

Look at eofiles $ cd eofiles $ less jobname. e 12345 $ less jobname. o 12345

Locate output files $ cd /Users/identikey/Day 4/QC $ ls Move html file to visualizer folder $ cp Example_X_fastqc. html / projects/sreadgroup/students/ identikey/ OR: Use File. Zilla to transfer html files to your own computer


Running Trimmomatic: Quick Start Settings $ java –jar /opt/trimmomatic/ 0. 32/trimmomatic-0. 32. jar $ cd /Users/identikey/PBS $ nano QS_trim. pbs

Replace anything with "<___>" with your desired value and "amso 5880" with your identikey • • Job name Queue eofiles Email

Trimmomatic Settings java -jar /opt/trimmomatic/0. 32/trimmomatic -0. 32. jar SE -threads 4 -phred 33 • • call program Single end setting Multithreading Phred 33 quality scores

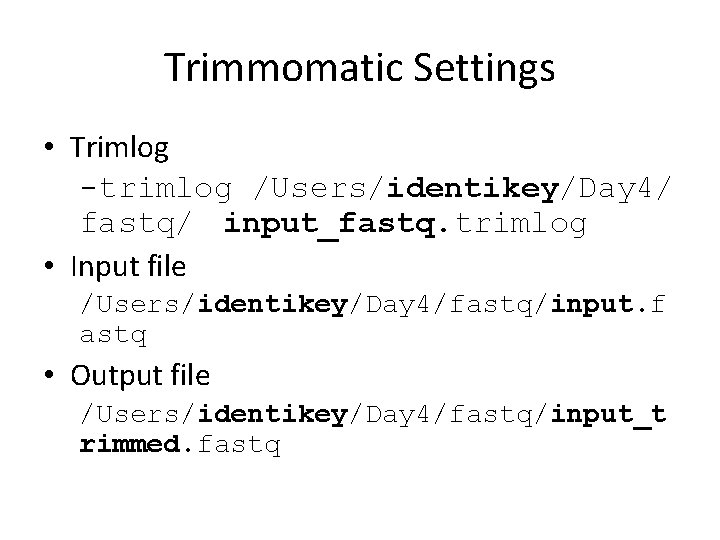
Trimmomatic Settings • Trimlog -trimlog /Users/identikey/Day 4/ fastq/ input_fastq. trimlog • Input file /Users/identikey/Day 4/fastq/input. f astq • Output file /Users/identikey/Day 4/fastq/input_t rimmed. fastq

Trimmomatic Settings ILLUMINACLIP: /opt/trimmomatic/0. 3 2/adaptors/Tru. Seq 3 -PE 2. fa: 2: 30: 10 LEADING: 3 TRAILING: 3 SLIDINGWINDOW: 4: 15 MINLEN: 36

Run trimmomatic & look at eofiles $ qsub QS_trim. pbs $ qstat –u identikey $ cd eofiles $ less job_name. e 12345 $ less job_name. o 12345

Check trimlog and fastq_trimmed files $ cd /Users/identikey/Day 4/fastq $ ls $ head -12 input. fastq $ head -12 input_trimmed. fastq $ less input_fastq. trimlog

Copy trimmomatic PBS to a new file $ cd. . /PBS $ ls $ cp QS_trim. pbs new_trim. pbs $ nano new_trim. pbs

Update new_trim. pbs • Job name • Settings java -jar /opt/trimmomatic/0. 32/ trimmomatic 0. 32. jar SE -threads 4 - phred 33 -trimlog /Users/amso 5880/Day 4/fastq/ input_fastq_cropped. trimlog input. fastq input_cropped. fastq CROP: 40 HEADCROP: 10

Run trimmomatic & look at eofiles $ qsub new_trim. pbs $ qstat –u identikey $ cd eofiles $ less new_job_name. e 12345 $ less new_job_name. o 12345

Check trimlog and fastq_trimmed files $ cd /Users/identikey/Day 4/fastq $ ls $ head -12 input. fastq $ head -12 input_cropped. fastq $ less input_fastq_cropped. trimlog