Sets Sets are lists with no duplicate entries

![Sets contd. a = set(["Jake", "John", "Eric"]) b = set(["John", "Jill"]) print a. symmetric_difference(b) Sets contd. a = set(["Jake", "John", "Eric"]) b = set(["John", "Jill"]) print a. symmetric_difference(b)](https://slidetodoc.com/presentation_image_h2/dfc99293389753e76663f01101b421c2/image-2.jpg)







- Slides: 16

Sets • Sets are lists with no duplicate entries. print set("my name is Eric and Eric is my name". split()) returns set [‘my’, name, ‘is’, ‘Eric’, ‘and’] • Sets are a powerful tool in Python since they have the ability to calculate differences and intersections between other sets. a = set(["Jake", "John", "Eric"]) b = set(["John", "Jill"]) a. intersection(b) b. intersection(a)
![Sets contd a setJake John Eric b setJohn Jill print a symmetricdifferenceb Sets contd. a = set(["Jake", "John", "Eric"]) b = set(["John", "Jill"]) print a. symmetric_difference(b)](https://slidetodoc.com/presentation_image_h2/dfc99293389753e76663f01101b421c2/image-2.jpg)
Sets contd. a = set(["Jake", "John", "Eric"]) b = set(["John", "Jill"]) print a. symmetric_difference(b) print b. symmetric_difference(a) print a. difference(b) print b. difference(a) print a. union(b)

Exception handling • Python's solution to errors are exceptions. You might have seen an exception before. def do_stuff_with_number(n): print n the_list = [1, 2, 3, 4, 5] for i in range(20): try: do_stuff_with_number(the_list[i]) except Index. Error: # Raised when accessing a non-existing index of a list do_stuff_with_number(0)

more information www. python. org

Bio. Python http: //biopython. org/wiki/Biopython Download http: //biopython. org/wiki/Download & Installation Documentation http: //biopython. org/wiki/Category%3 AWiki_Documentation

Bio. Python Key features: • • Sequences Sequence Annotation I/O Operations Accessing online databases Multiple sequence alignments BLAST and many more …

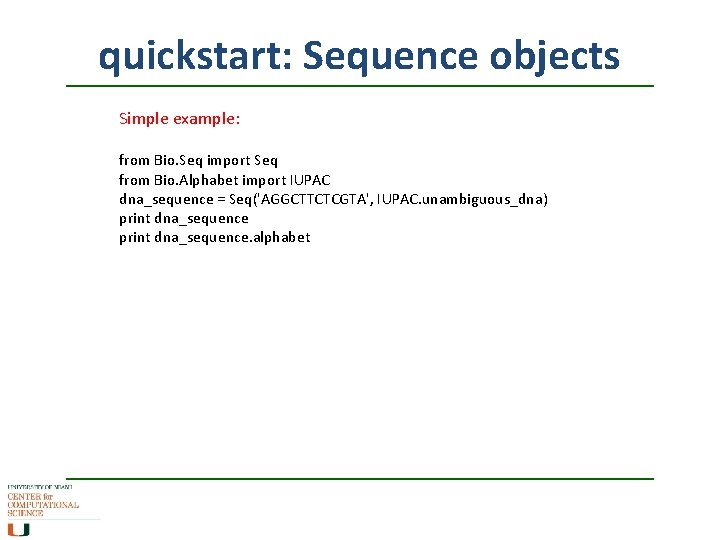
quickstart: Sequence objects Simple example: from Bio. Seq import Seq from Bio. Alphabet import IUPAC dna_sequence = Seq('AGGCTTCTCGTA', IUPAC. unambiguous_dna) print dna_sequence. alphabet

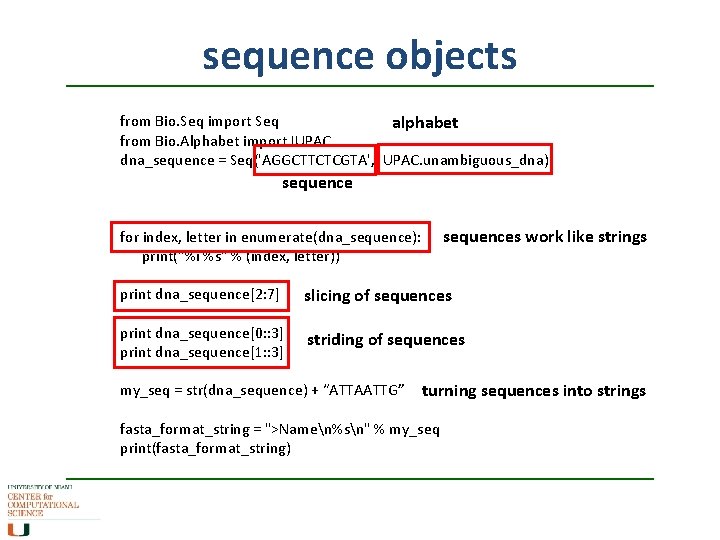
sequence objects from Bio. Seq import Seq alphabet from Bio. Alphabet import IUPAC dna_sequence = Seq('AGGCTTCTCGTA', IUPAC. unambiguous_dna) sequences work like strings for index, letter in enumerate(dna_sequence): print("%i %s" % (index, letter)) print dna_sequence[2: 7] slicing of sequences print dna_sequence[0: : 3] print dna_sequence[1: : 3] striding of sequences my_seq = str(dna_sequence) + “ATTAATTG” turning sequences into strings fasta_format_string = ">Namen%sn" % my_seq print(fasta_format_string)

sequence objects from Bio. Seq import Seq from Bio. Alphabet import IUPAC my_seq = Seq("GATCGATGGGCCTATATAGGATCGAAAATCGC”, IUPAC. unambiguous_dna) print my_seq. complement() print my_seq. reverse_complement() messenger_rna = my_seq. transcribe() print messenger_rna making complements making m. RNA

sequence objects from Bio. Seq import Seq from Bio. Alphabet import IUPAC messenger_rna = Seq("AUGGCCAUUGUAAUGGGCCGCUGAAAGGGUGCCCGAUAG”, IUPAC. unambiguous_rna) print messenger_rna. translate() translation coding_dna = Seq("ATGGCCATTGTAATGGGCCGCTGAAAGGGTGCCCGATAG", IUPAC. unambiguous_dna) translation print coding_dna. translate() print coding_dna. translate(table = 2) translation using different tables #translation tables https: //www. ncbi. nlm. nih. gov/Taxonomy/Utils/wprintgc. cgi

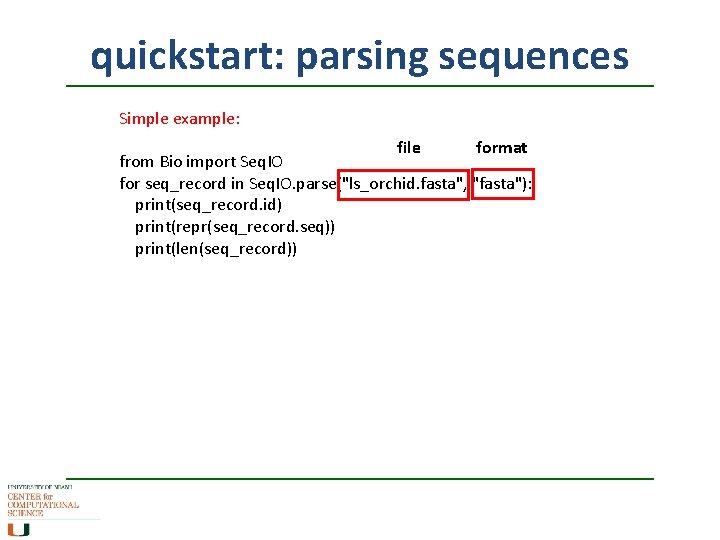
quickstart: parsing sequences Simple example: file format from Bio import Seq. IO for seq_record in Seq. IO. parse("ls_orchid. fasta", "fasta"): print(seq_record. id) print(repr(seq_record. seq)) print(len(seq_record))

seq. Record object. sequence itself, typically a Seq object. . id primary id, string. name common name, string. description human readable description, string. letter_annotations Holds per-letter-annotations using a (restricted) dictionary of additional information, Python sequence. annotations additional information, dictionary. features A list of Seq. Feature objects with more structured information about the features on a sequence (e. g. position of genes on a genome, or domains on a protein sequence). dbxrefs database cross-references, string

seq. Record object from scratch from Bio. Seq import Seq simple_seq = Seq("GATC") from Bio. Seq. Record import Seq. Record simple_seq_r = Seq. Record(simple_seq) simple_seq_r. id = (“ 0001”) simple_seq_r. name = (“MFG 1”) simple_seq_r. description = "Made up sequence” print simple_seq_r reading the information from Bio import Seq. IO record = Seq. IO. read("NC_005816. fna", "fasta") print record

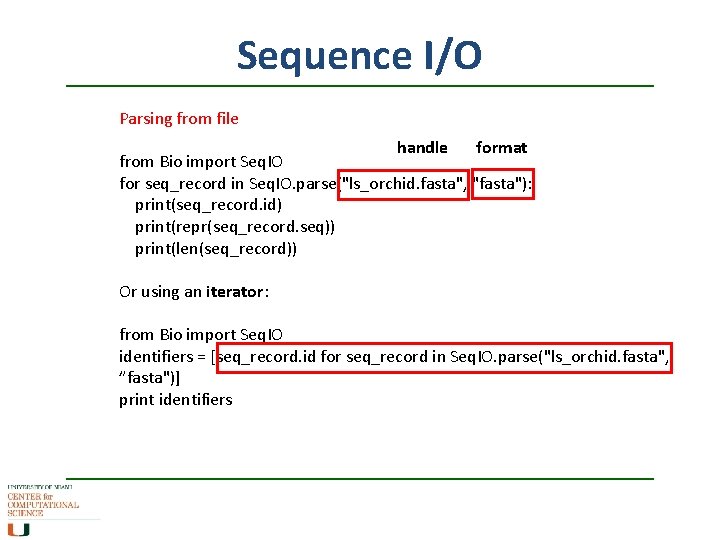
Sequence I/O Parsing from file handle format from Bio import Seq. IO for seq_record in Seq. IO. parse("ls_orchid. fasta", "fasta"): print(seq_record. id) print(repr(seq_record. seq)) print(len(seq_record)) Or using an iterator: from Bio import Seq. IO identifiers = [seq_record. id for seq_record in Seq. IO. parse("ls_orchid. fasta", ”fasta")] print identifiers

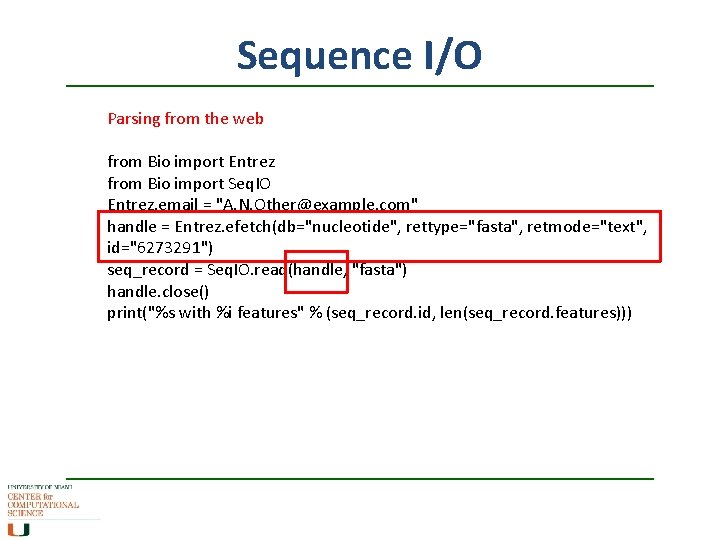
Sequence I/O Parsing from the web from Bio import Entrez from Bio import Seq. IO Entrez. email = "A. N. Other@example. com" handle = Entrez. efetch(db="nucleotide", rettype="fasta", retmode="text", id="6273291") seq_record = Seq. IO. read(handle, "fasta") handle. close() print("%s with %i features" % (seq_record. id, len(seq_record. features)))

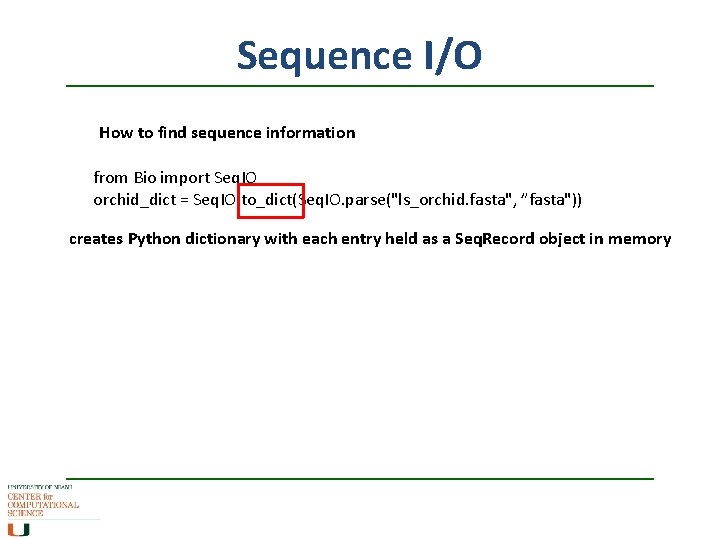
Sequence I/O How to find sequence information from Bio import Seq. IO orchid_dict = Seq. IO. to_dict(Seq. IO. parse("ls_orchid. fasta", ”fasta")) creates Python dictionary with each entry held as a Seq. Record object in memory