Set Up a Working Environment Python help command












- Slides: 13

Set Up a Working Environment • Python help command • pydoc server • Argon simulation 1. 2. 3. 4. Run simulation Visualize trajectory Modify script Repeat • Water simulation Run, Visualize, Modify, Repeat 1

Python’s help Command From the Python interpreter type: >>> >>> >>> import simtk. unit as unit help(unit) help(unit. calorie) help(unit. Quantity. value_in_unit) Warning: Never run from the Py. Open. MM install directory because it will load the wrong modules. 2

Python’s help Command From the Python interpreter type: >>> >>> >>> import simtk. chem. openmm as mm help(mm) help(mm. System. add. Force) help(mm. Nonbonded. Force) help(mm. Verlet. Integrator. step) help(mm. Context. set. Positions) help(mm. Context. get. State) help(mm. state. get. Potential. Energy) 3

Browser Access using pydoc Mac and Linux: From a command prompt, type the following: $ pydoc -p 8888 (maybe “pydoc 25 –p 8888” on snow leopard) Windows: From a command prompt, type the following: > C: Python 26python. exe Python 26Libpydoc. py -p 8888 For all Sim. TK packages, point your browser to: http: //localhost: 8888/simtk Best place to look for Open. MM classes is: http: //localhost: 8888/simtk. chem. openmm 4

pydoc for System Class (in simtk. chem. openmm) 5

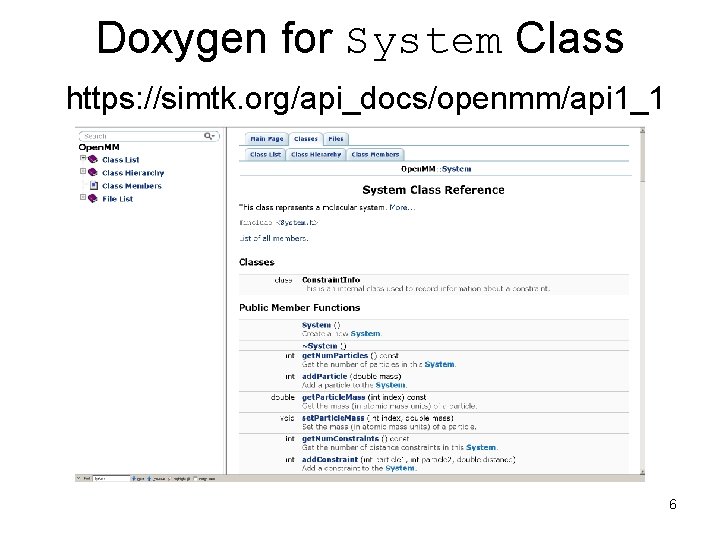
Doxygen for System Class https: //simtk. org/api_docs/openmm/api 1_1 6

Run Argon Simulation A 500 femtosecond, three argon atom, constant energy simulation. Atoms interact with a Lennard-Jones potential and a very large cutoff distance. Integration is done with Verlet integrator. Center of mass motion is periodically removed to make it easier to visualize the system. Mac and Linux: $ cd examples/argon $ python run. Argon. py argon. pdb Windows: > cd examplesargon > C: Python 26python run. Argon. py argon. pdb 7

Visualize Argon Simulation Visualize the result using VMD (http: //www. ks. uiuc. edu/Research/vmd) 1. Start VMD 2. Open argon. pdb (File -> New Molecule) 3. Show VDW surface (Graphics -> Representations and then change the Drawing Method to VDW) 4. Replay the trajectory by pushing the play button in the lower right corner of the “VMD Main” windows 8

Modified Argon Simulation Edit run. Argon. py and rerun the simulation. 1. Add more argon atoms (~20) and increase simulation length (~20 ps). Starting atom positions can be straight lines, circle, etc). 2. Instead of constant energy, run a constant temperature simulation using the Andersen thermostat (which is a Force subclass). The collision frequency can be in the range 0. 001 to 0. 1 reciprocal picoseconds. 3. Add harmonic bond forces between argon atoms to make a coarsegrained polymer model. Parameters are up to you, but force constants of about 100 (kcal/mol)/Å^2 should work. Remember to add nonbond exceptions using the Nonbonded. Force. add. Exception method--for charge. Prod, sigma and epsilon, use 0, 1, 0 respectively. 4. Time permitting, add harmonic angle and periodic torsion forces as well with force constants of about 10 (kcal/mol)/(radian^2) and 1 kcal/mol. 9

Water Simulation A 4 picosecond, 53 TIP 3 P water molecules, constant temperature (300 K) simulation. Integration is done with Verlet integrator and center of mass motion is removed at each step to simplify visualizing the trajectory. Also, there is a harmonic forces between each Oxygen atom pair Mac and Linux: $ cd examples/H 2 O $ python run. H 2 O. py wat 53. pdb H 2 O. pdb Windows: > cd examplesH 2 O > C: Python 26python run. H 2 O. py wat 53. pdb H 2 O. pdb 10

Modified Water Simulation Edit run. H 20. py and rerunning the simulation. Try the following: 1. Try a different integrator, such as Langevin (remember to remove Andersen thermostat). Try different temperatures, including 0 K (can be used as a minimize). 2. Change restraint between oxygen atoms to a custom Nonbonded force restraint using the Custom. Nonbonded. Force class. A possible energy equation is “K 1*K 2*(r/(R 1+R 2))^4”, but use what you want (note the change is simulation time). 11

Other Examples • For example code showing how to construct various systems, see some of John Chodera’s test code: simtk/chem/openmm/extras/testsystems. py • More examples will be added before the next release. 12

Help and Feedback From Py. Open. MM Home Page: https: //simtk. org/home/pyopenmm 1. Forums: Advanced -> Public Forums 2. Bug Tracking: Advanced -> Features & Bugs Email Addresses: Randy Radmer: radmer@stanford. edu Chris Bruns: cmbruns@stanford. edu 13