Sequencing insert 2 Parts 1 Preparing ladder ending















- Slides: 68

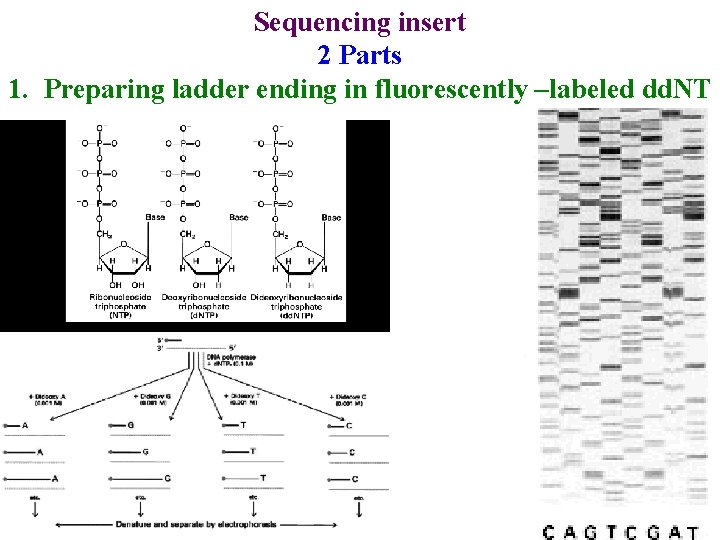
Sequencing insert 2 Parts 1. Preparing ladder ending in fluorescently –labeled dd. NT 1/ Mega. BACE 4500 Training Module 1 / 1/10/2022

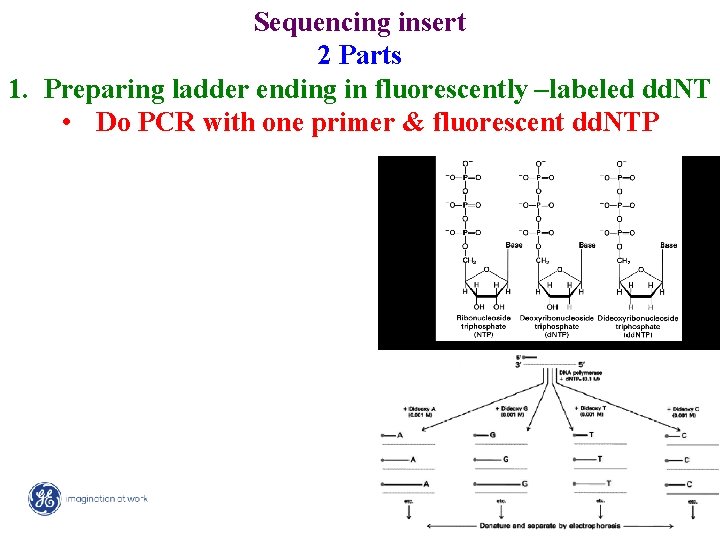
Sequencing insert 2 Parts 1. Preparing ladder ending in fluorescently –labeled dd. NT • Do PCR with one primer & fluorescent dd. NTP 2/ Mega. BACE 4500 Training Module 1 / 1/10/2022

1. Preparing ladder ending in fluorescently –labeled dd. NT • Do PCR with one primer & fluorescent dd. NTP • Excitation transfer: Fluorescein absorbs 488 nm from argon laser • Transfers energy to acceptor dye which fluoresces • dd. G, dd. A, dd. T & dd. C, have different acceptor dyes which fluoresce different colors 3/ Mega. BACE 4500 Training Module 1 / 1/10/2022

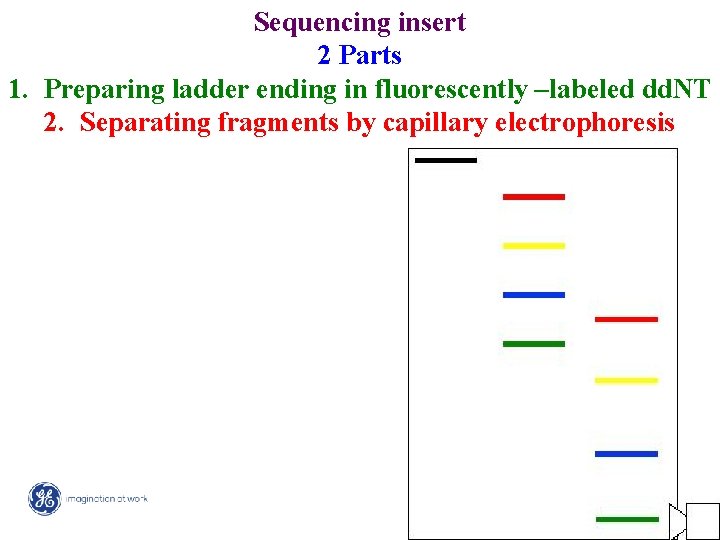
Sequencing insert 2 Parts 1. Preparing ladder ending in fluorescently –labeled dd. NT 2. Separating fragments by capillary electrophoresis 4/ Mega. BACE 4500 Training Module 1 / 1/10/2022

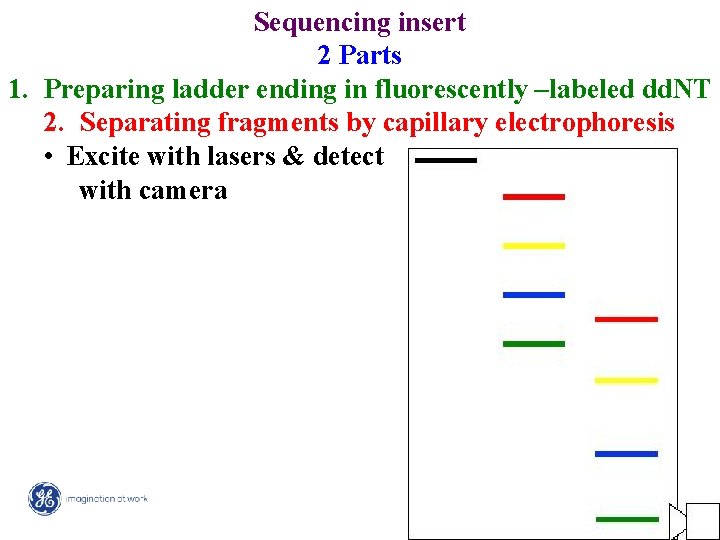
Sequencing insert 2 Parts 1. Preparing ladder ending in fluorescently –labeled dd. NT 2. Separating fragments by capillary electrophoresis • Excite with lasers & detect with camera 5/ Mega. BACE 4500 Training Module 1 / 1/10/2022

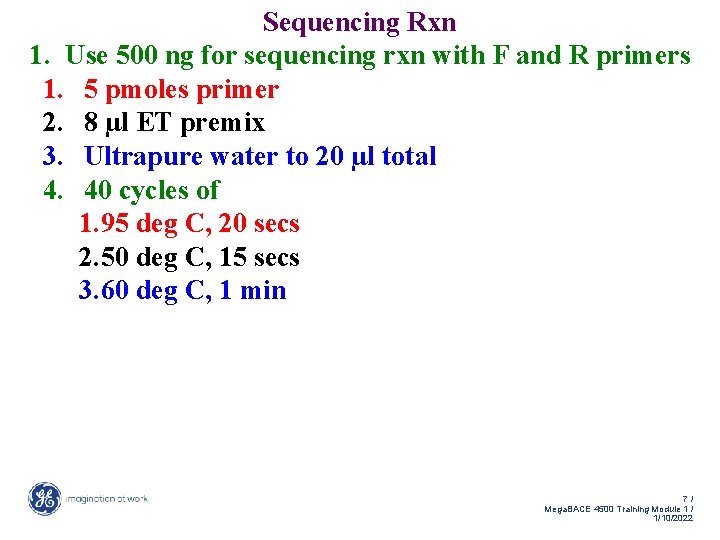
Sequencing Rxn 1. Use 500 ng for sequencing rxn with F and R primers 1. 5 pmoles primer 2. 8 µl ET premix 3. Ultrapure water to 20 µl total 6/ Mega. BACE 4500 Training Module 1 / 1/10/2022

Sequencing Rxn 1. Use 500 ng for sequencing rxn with F and R primers 1. 5 pmoles primer 2. 8 µl ET premix 3. Ultrapure water to 20 µl total 4. 40 cycles of 1. 95 deg C, 20 secs 2. 50 deg C, 15 secs 3. 60 deg C, 1 min 7/ Mega. BACE 4500 Training Module 1 / 1/10/2022

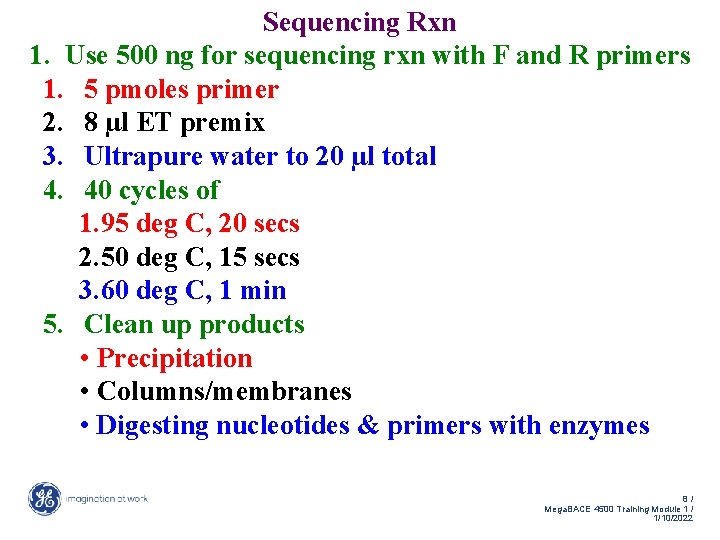
Sequencing Rxn 1. Use 500 ng for sequencing rxn with F and R primers 1. 5 pmoles primer 2. 8 µl ET premix 3. Ultrapure water to 20 µl total 4. 40 cycles of 1. 95 deg C, 20 secs 2. 50 deg C, 15 secs 3. 60 deg C, 1 min 5. Clean up products • Precipitation • Columns/membranes • Digesting nucleotides & primers with enzymes 8/ Mega. BACE 4500 Training Module 1 / 1/10/2022

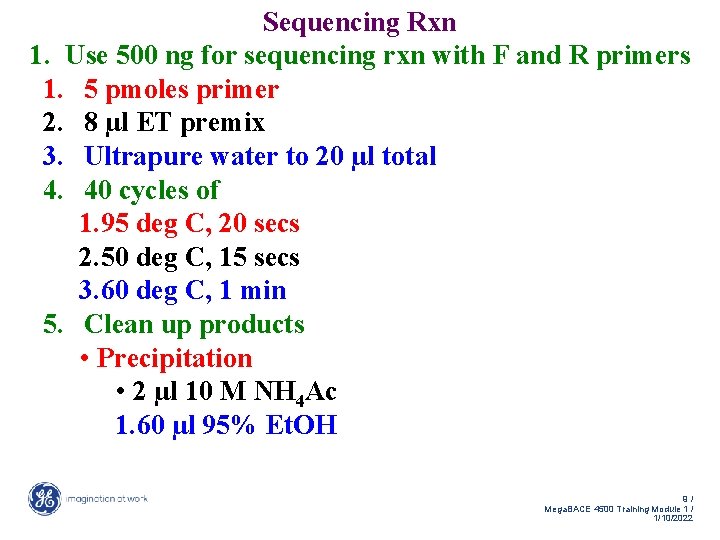
Sequencing Rxn 1. Use 500 ng for sequencing rxn with F and R primers 1. 5 pmoles primer 2. 8 µl ET premix 3. Ultrapure water to 20 µl total 4. 40 cycles of 1. 95 deg C, 20 secs 2. 50 deg C, 15 secs 3. 60 deg C, 1 min 5. Clean up products • Precipitation • 2 µl 10 M NH 4 Ac 1. 60 µl 95% Et. OH 9/ Mega. BACE 4500 Training Module 1 / 1/10/2022

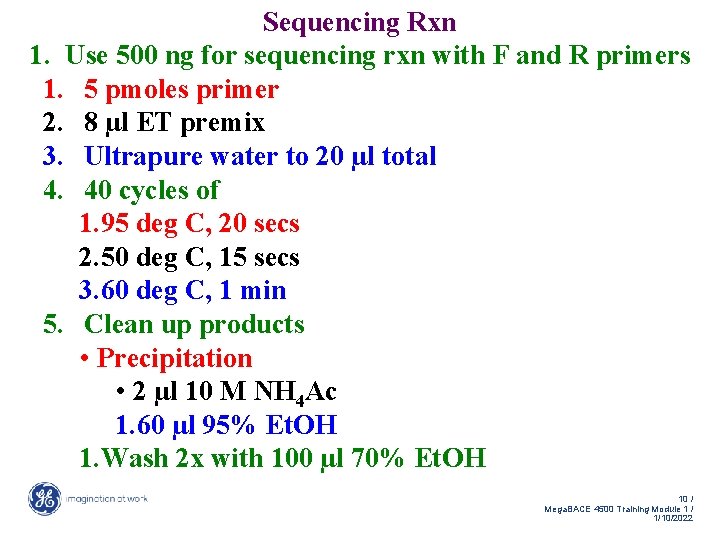
Sequencing Rxn 1. Use 500 ng for sequencing rxn with F and R primers 1. 5 pmoles primer 2. 8 µl ET premix 3. Ultrapure water to 20 µl total 4. 40 cycles of 1. 95 deg C, 20 secs 2. 50 deg C, 15 secs 3. 60 deg C, 1 min 5. Clean up products • Precipitation • 2 µl 10 M NH 4 Ac 1. 60 µl 95% Et. OH 1. Wash 2 x with 100 µl 70% Et. OH 10 / Mega. BACE 4500 Training Module 1 / 1/10/2022

Sequencing Rxn 1. Use 500 ng for sequencing rxn with F and R primers 1. 5 pmoles primer 2. 8 µl ET premix 3. Ultrapure water to 20 µl total 4. 40 cycles of 1. 95 deg C, 20 secs 2. 50 deg C, 15 secs 3. 60 deg C, 1 min 5. Clean up products • Precipitation • 2 µl 10 M NH 4 Ac 1. 60 µl 95% Et. OH 1. Wash 2 x with 100 µl 70% Et. OH 2. Air-dry, dissolve in 10 µl Sequencing loading buffer 11 / Mega. BACE 4500 Training Module 1 / 1/10/2022

Fragment separation 1. load capillaries with polyacrylamide @ 2000 psi 12 / Mega. BACE 4500 Training Module 1 / 1/10/2022

Fragment separation 1. load capillaries with polyacrylamide @ 2000 psi 2. Prep capillaries for sample 13 / Mega. BACE 4500 Training Module 1 / 1/10/2022

Fragment separation 1. load capillaries with polyacrylamide @ 2000 psi 2. Prep capillaries for sample 3. “Electroinject” the samples • Apply 3000 volts for 60 seconds 14 / Mega. BACE 4500 Training Module 1 / 1/10/2022

Fragment separation 1. load capillaries with polyacrylamide @ 2000 psi 2. Prep capillaries for sample 3. “Electroinject” the samples • Apply 3000 volts for 60 seconds • Only removes electrolytes: reduces [], not volume! 15 / Mega. BACE 4500 Training Module 1 / 1/10/2022

1. 2. 3. 4. Fragment separation load capillaries with polyacrylamide @ 2000 psi Prep capillaries for sample “Electroinject” the samples • Apply 3000 volts for 60 seconds • Only removes electrolytes: reduces [], not volume! Switch to “run’: standard = 100 minutes @ 9000 volts 16 / Mega. BACE 4500 Training Module 1 / 1/10/2022

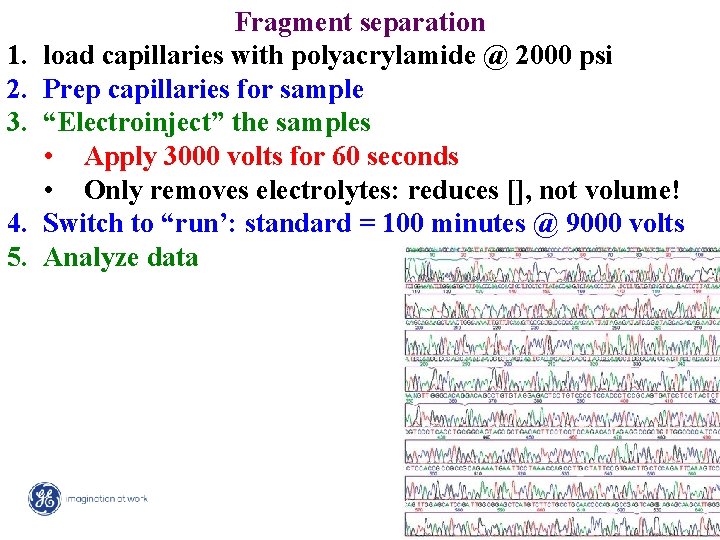
1. 2. 3. 4. 5. Fragment separation load capillaries with polyacrylamide @ 2000 psi Prep capillaries for sample “Electroinject” the samples • Apply 3000 volts for 60 seconds • Only removes electrolytes: reduces [], not volume! Switch to “run’: standard = 100 minutes @ 9000 volts Analyze data 17 / Mega. BACE 4500 Training Module 1 / 1/10/2022

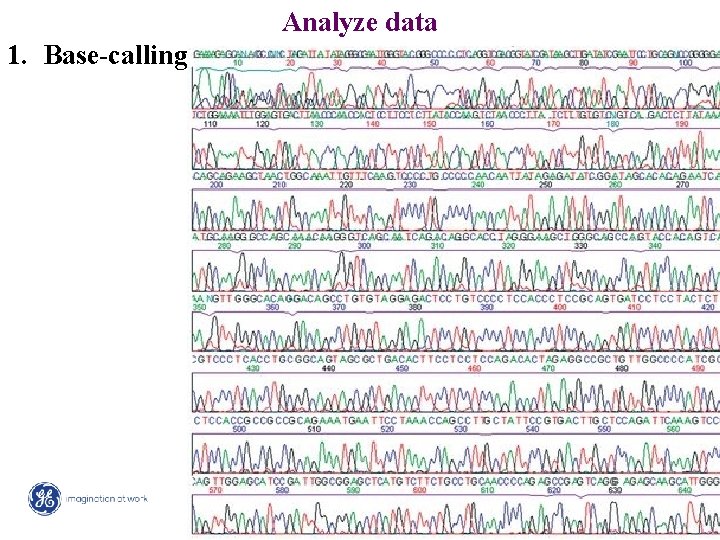
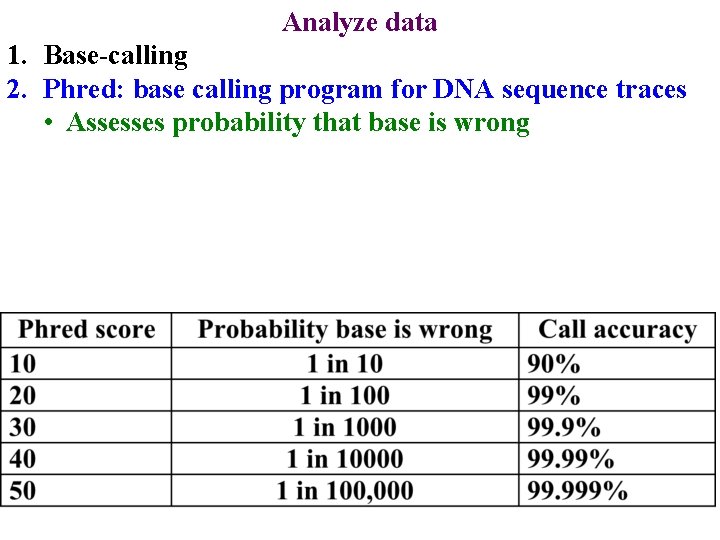
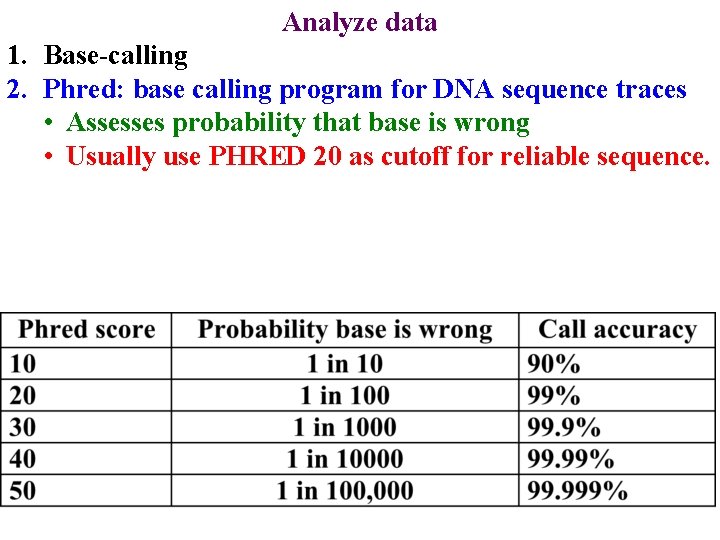
Analyze data 1. Base-calling 18 / Mega. BACE 4500 Training Module 1 / 1/10/2022

Analyze data 1. Base-calling 2. Phred: base calling program for DNA sequence traces • Assesses probability that base is wrong 19 / Mega. BACE 4500 Training Module 1 / 1/10/2022

Analyze data 1. Base-calling 2. Phred: base calling program for DNA sequence traces • Assesses probability that base is wrong • Usually use PHRED 20 as cutoff for reliable sequence. 20 / Mega. BACE 4500 Training Module 1 / 1/10/2022

Mega. BACE 500 DNA Analysis System overview and training Michael Reagin Applications Consultant Genomics Applications

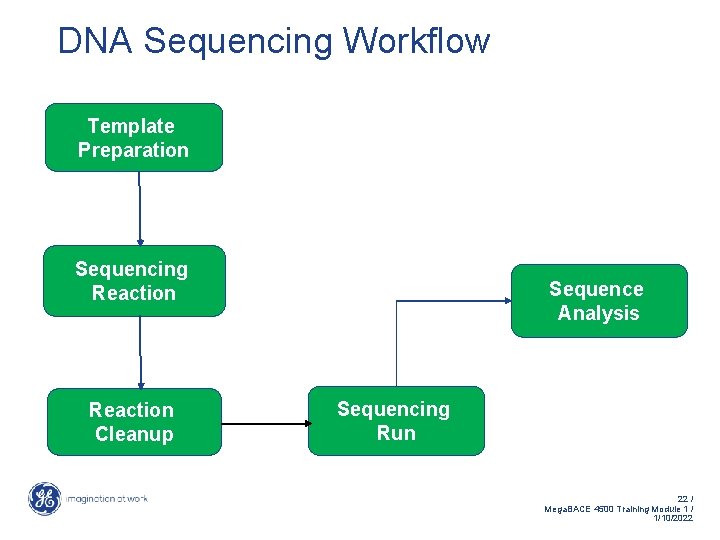
DNA Sequencing Workflow Template Preparation Sequencing Reaction Cleanup Sequence Analysis Sequencing Run 22 / Mega. BACE 4500 Training Module 1 / 1/10/2022

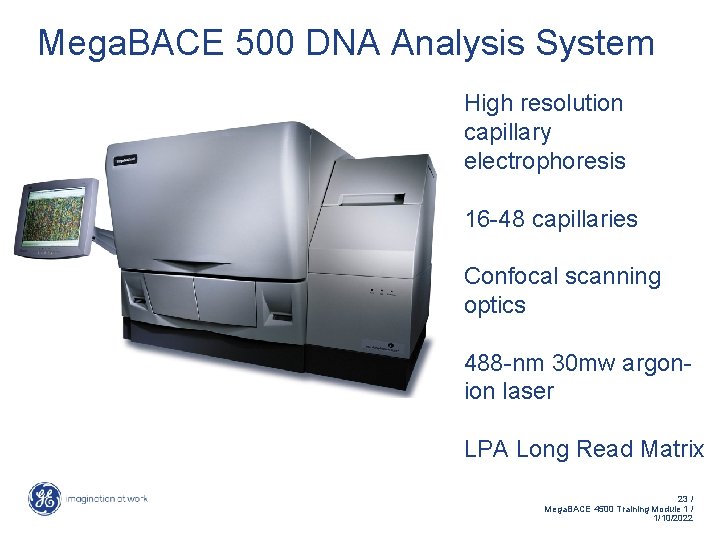
Mega. BACE 500 DNA Analysis System High resolution capillary electrophoresis 16 -48 capillaries Confocal scanning optics 488 -nm 30 mw argonion laser LPA Long Read Matrix 23 / Mega. BACE 4500 Training Module 1 / 1/10/2022

Mega. BACE DNA Analysis System Pneumatics Low pressure N 2 (100 psi) – Move parts within the machine – Fill the capillaries with water High pressure N 2 (1000 psi) – Fill the capillaries with LPA matrix – Flush LPA matrix from the caps 24 / Mega. BACE 4500 Training Module 1 / 1/10/2022

Mega. BACE Workflow • Inject Linear Polyacrylamide matrix (LPA) into caps. • Allow matrix to equilibrate 1 - 5 min. • Pre-run machine for 5 min. • Electro-kinetically inject labeled DNA 10 - 60 secs. • Use electric current to move DNA through capillaries (75 min. to 5 hrs. ) • DNA migrates past excitation/detection window, where fluorescent dyes are excited by laser, and fluorescence is directed back into machine for detection • Fluorescent data is collected, automatically analyzed, and exported to different file formats (if needed) 25 / Mega. BACE 4500 Training Module 1 / 1/10/2022

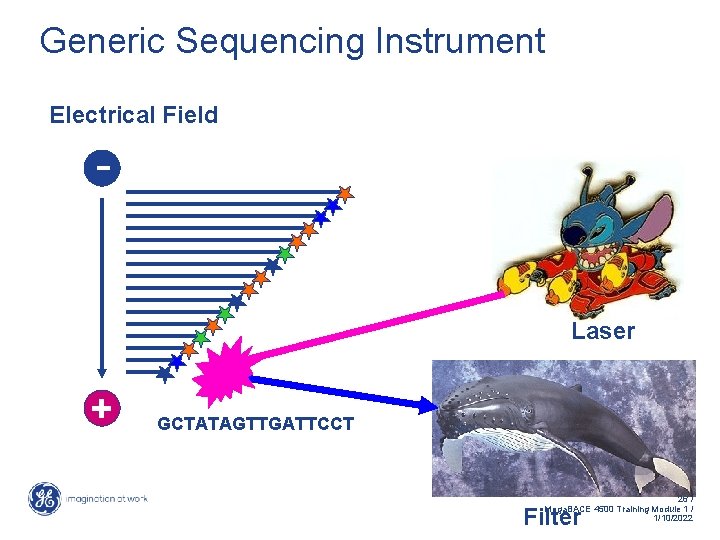
Generic Sequencing Instrument Electrical Field - Laser + GCTATAGTTGATTCCT 26 / Mega. BACE 4500 Training Module 1 / 1/10/2022 Filter

Capillary Electrophoresis is the size separation of DNA fragments accomplished by applying a potential across a gel-filled, µm-sized, narrow-bore capillary. CE separations yield fast and high-resolution separations A variety of detection modes is possible Capillaries may be re-used due to the properties of the Linear Polymer Matrix Minimal manpower and lower overhead for HT facilities. 27 / Mega. BACE 4500 Training Module 1 / 1/10/2022

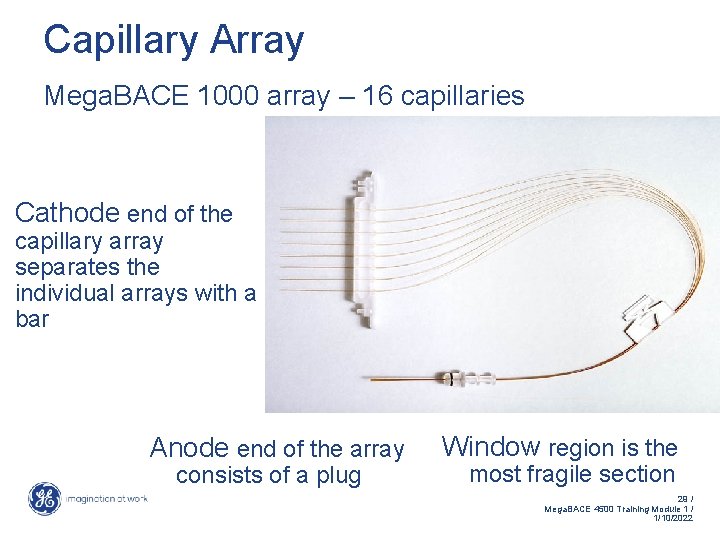
Capillary Array The capillary acts as a container for the matrix, as a voltage conduit, and as an optical lens. Supplied in pre-assembled arrays of 16 individual capillaries Physical properties: • Outer diatmeter: 200µm • Inner diameter: 75µm • Total length: 62. 5 cm • Effective length: 40 cm Narrow-bore capillaries providing efficient heat dissipation (Joule heating) • Diffusion reduced to narrow-bore of capillaries 28 / Mega. BACE 4500 Training Module 1 / 1/10/2022

Capillary Array Mega. BACE 1000 array – 16 capillaries Cathode end of the capillary array separates the individual arrays with a bar Anode end of the array consists of a plug Window region is the most fragile section 29 / Mega. BACE 4500 Training Module 1 / 1/10/2022

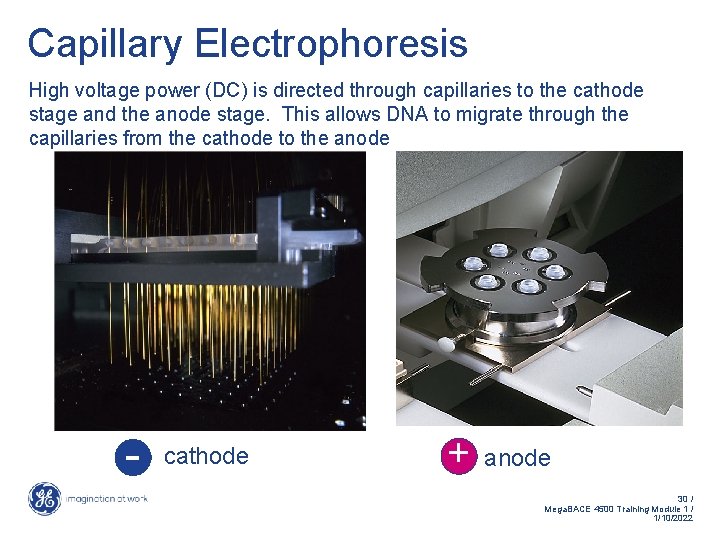
Capillary Electrophoresis High voltage power (DC) is directed through capillaries to the cathode stage and the anode stage. This allows DNA to migrate through the capillaries from the cathode to the anode - cathode + anode 30 / Mega. BACE 4500 Training Module 1 / 1/10/2022

Mega. BACE 1000 CE System - Cathode Stage + Anode Stage 31 / Mega. BACE 4500 Training Module 1 / 1/10/2022

Linear Poly. Acrylamide (LPA) LPA sequencing matrix dynamically coats capillary walls • Improves run-to-run consistency Reduced LPA concentration • Improves separation of longer DNA fragments Optimized formulation • Reduces matrix lot-to-lot variability • Improves manufacturing tolerances 32 / Mega. BACE 4500 Training Module 1 / 1/10/2022

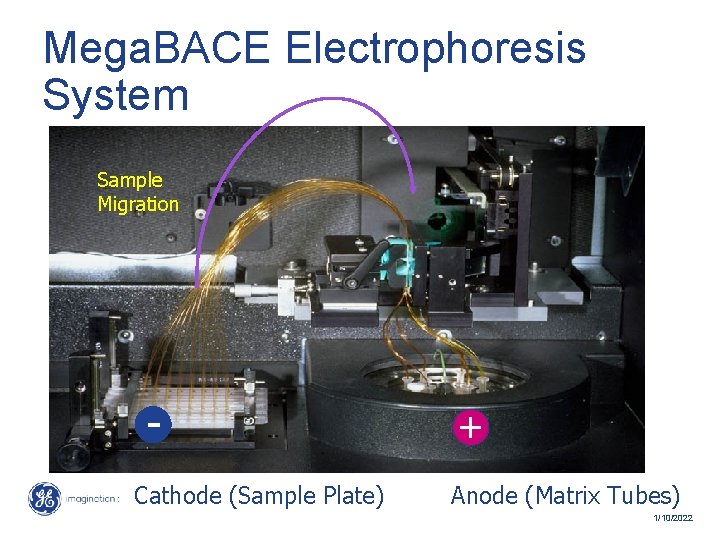
Mega. BACE Electrophoresis System Sample Migration Cathode (Sample Plate) + Anode (Matrix Tubes) 33 / Mega. BACE 4500 Training Module 1 / 1/10/2022

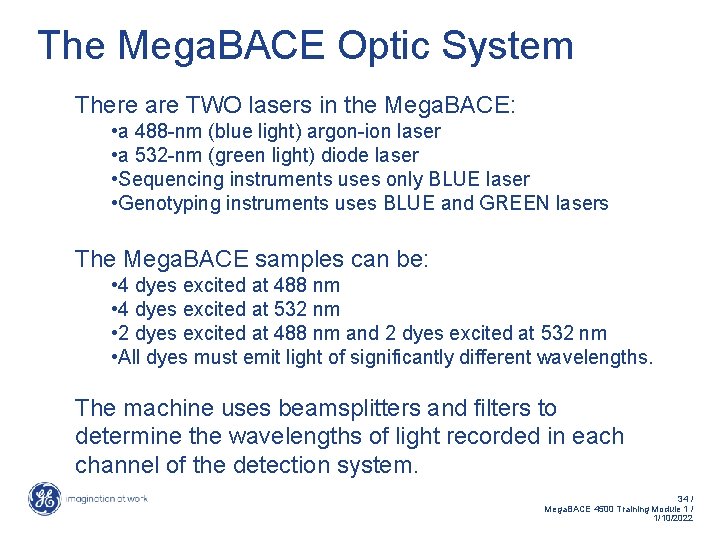
The Mega. BACE Optic System There are TWO lasers in the Mega. BACE: • a 488 -nm (blue light) argon-ion laser • a 532 -nm (green light) diode laser • Sequencing instruments uses only BLUE laser • Genotyping instruments uses BLUE and GREEN lasers The Mega. BACE samples can be: • 4 dyes excited at 488 nm • 4 dyes excited at 532 nm • 2 dyes excited at 488 nm and 2 dyes excited at 532 nm • All dyes must emit light of significantly different wavelengths. The machine uses beamsplitters and filters to determine the wavelengths of light recorded in each channel of the detection system. 34 / Mega. BACE 4500 Training Module 1 / 1/10/2022

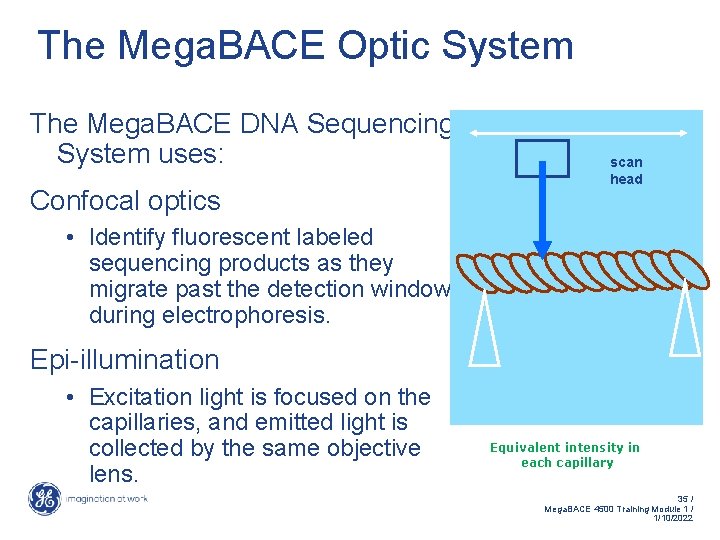
The Mega. BACE Optic System The Mega. BACE DNA Sequencing System uses: Confocal optics scan head • Identify fluorescent labeled sequencing products as they migrate past the detection window during electrophoresis. Epi-illumination • Excitation light is focused on the capillaries, and emitted light is collected by the same objective lens. Equivalent intensity in each capillary 35 / Mega. BACE 4500 Training Module 1 / 1/10/2022

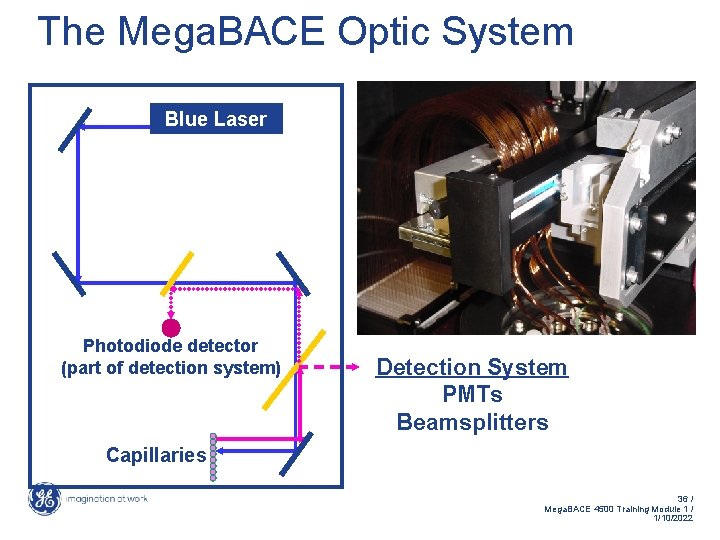
The Mega. BACE Optic System Blue Laser Photodiode detector (part of detection system) Detection System PMTs Beamsplitters Capillaries 36 / Mega. BACE 4500 Training Module 1 / 1/10/2022

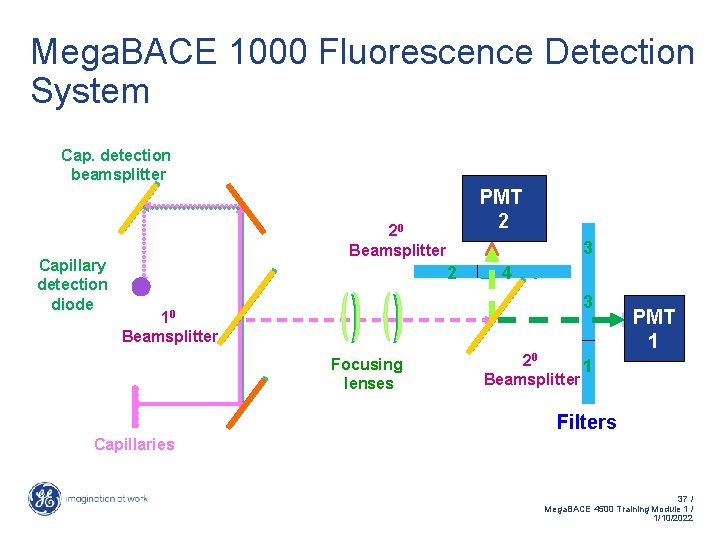
Mega. BACE 1000 Fluorescence Detection System Cap. detection beamsplitter Capillary detection diode PMT 2 20 Beamsplitter 3 2 2 4 4 3 1 10 Beamsplitter Focusing lenses 20 1 Beamsplitter PMT 1 Filters Capillaries 37 / Mega. BACE 4500 Training Module 1 / 1/10/2022

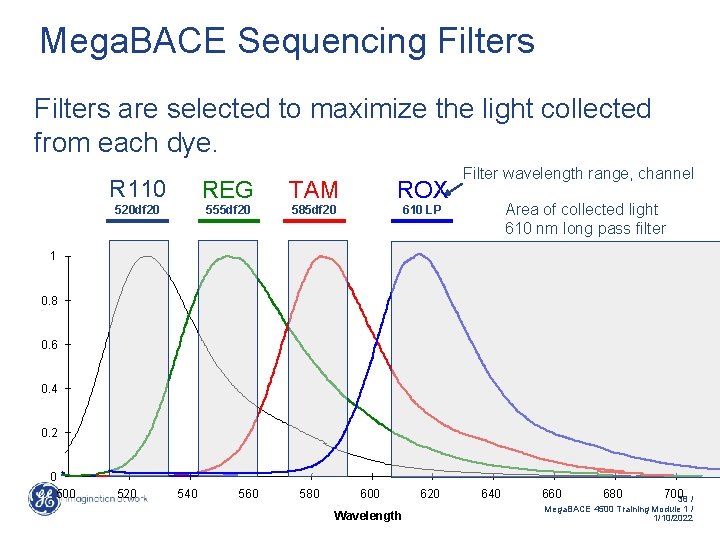
Mega. BACE Sequencing Filters are selected to maximize the light collected from each dye. R 110 REG TAM 520 df 20 555 df 20 585 df 20 ROX Filter wavelength range, channel Area of collected light 610 nm long pass filter 610 LP 1 0. 8 0. 6 0. 4 0. 2 0 500 520 540 560 580 600 Wavelength 620 640 660 680 70038 / Mega. BACE 4500 Training Module 1 / 1/10/2022

Host Scan Controller The Mega. BACE instrument is a SCSI device • The Mega. BACE system MUST be powered on BEFORE the computer workstation • “beeps” The Host Scan Controller (HSC) is the interface program between the computer and the instrument • The HSC must be opened before the ICM or the ICS • The HSC must be properly communicating 39 / Mega. BACE 4500 Training Module 1 / 1/10/2022

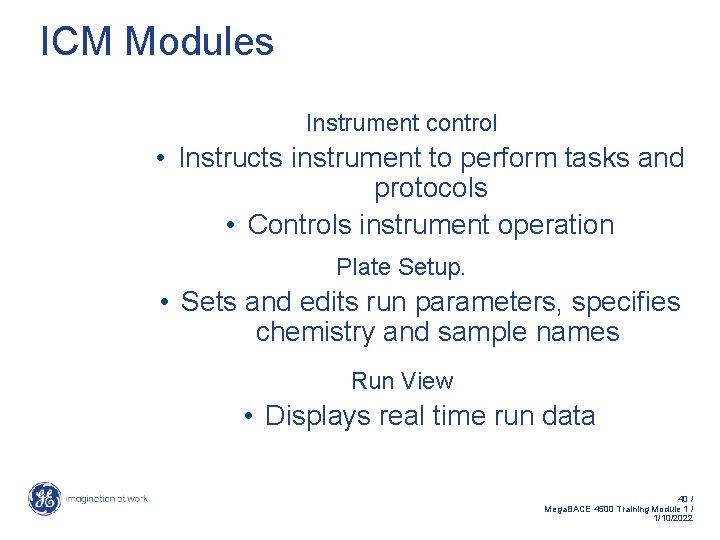
ICM Modules Instrument control • Instructs instrument to perform tasks and protocols • Controls instrument operation Plate Setup. • Sets and edits run parameters, specifies chemistry and sample names Run View • Displays real time run data 40 / Mega. BACE 4500 Training Module 1 / 1/10/2022

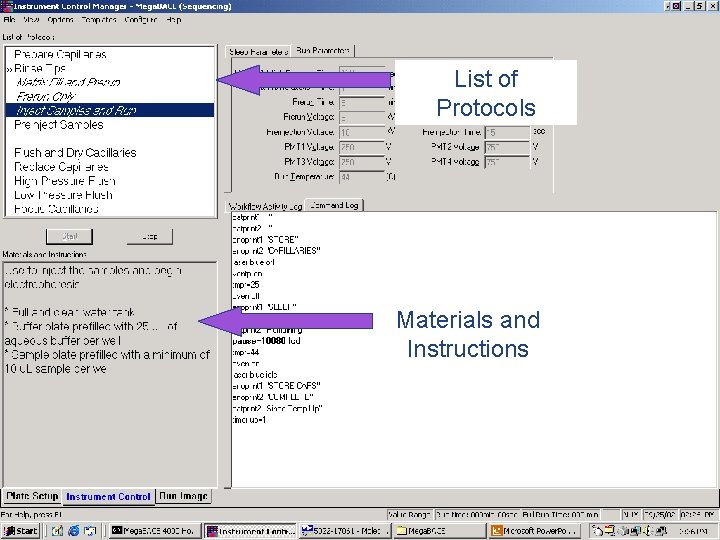
List of Protocols Materials and Instructions 41 / Mega. BACE 4500 Training Module 1 / 1/10/2022

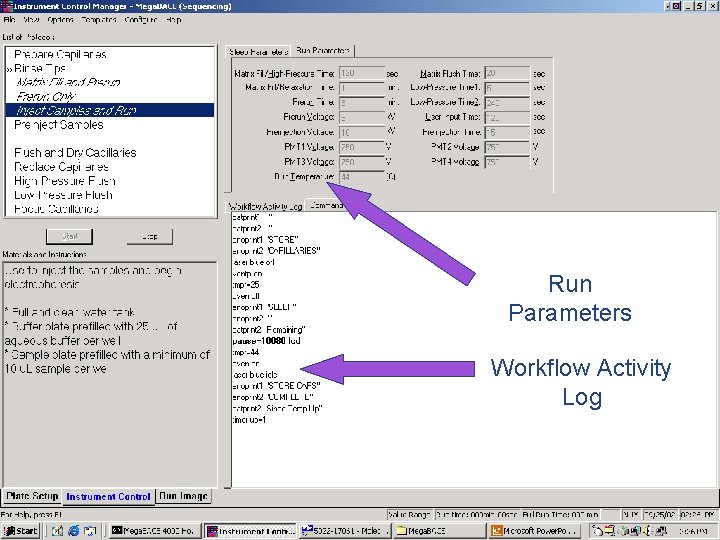
Run Parameters Workflow Activity Log 42 / Mega. BACE 4500 Training Module 1 / 1/10/2022

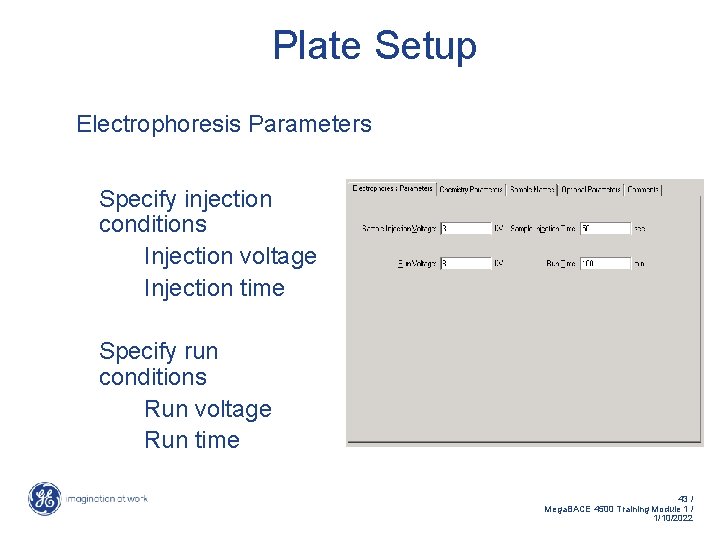
Plate Setup Electrophoresis Parameters Specify injection conditions Injection voltage Injection time Specify run conditions Run voltage Run time 43 / Mega. BACE 4500 Training Module 1 / 1/10/2022

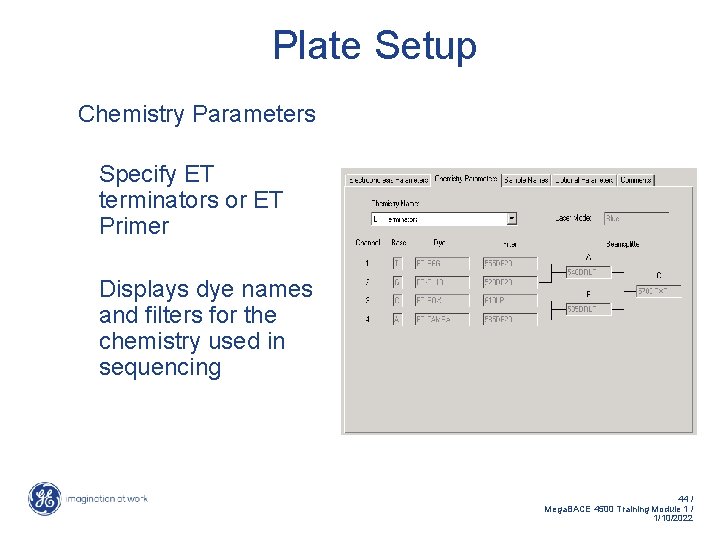
Plate Setup Chemistry Parameters Specify ET terminators or ET Primer Displays dye names and filters for the chemistry used in sequencing 44 / Mega. BACE 4500 Training Module 1 / 1/10/2022

Plate Setup Optional Parameters Specify Basecaller Adjust PMT sensitivity Change Run Temperature 45 / Mega. BACE 4500 Training Module 1 / 1/10/2022

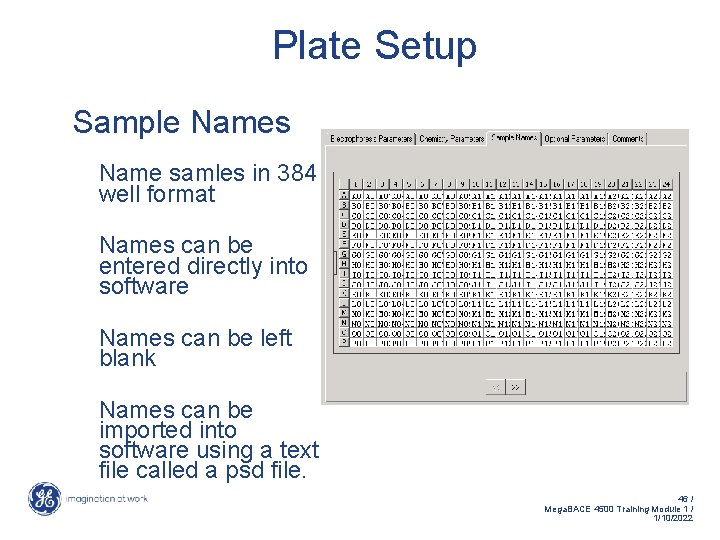
Plate Setup Sample Names Name samles in 384 well format Names can be entered directly into software Names can be left blank Names can be imported into software using a text file called a psd file. 46 / Mega. BACE 4500 Training Module 1 / 1/10/2022

ICM Protocols Mega. BACE Protocols are all based on the following: • Raising and Lowering the Cathode and Anode. • Applying Pressure to the Anode (30/1000 psi). • Applying Voltage across the Anode and Cathode. Mega. BACE Protocols all use the following: • Sample and Buffer Trays on the Cathode. • Water and Empty Tanks on the Cathode. • Water, LPA, & Focusing Dye in the Anode Tubes. 47 / Mega. BACE 4500 Training Module 1 / 1/10/2022

Instrument Run Protocols Rinse Tips • Performed only before the first run of the day • Cleans the tips of the capillaries before sample injection Matrix Fill and Prerun • LPA injected into capillaries from anode stage • matrix equilibration and prerun Inject Samples and Run • Sample electro-kinetically injected from cathode stage • Use electric current to move sample through capillaries for data collection. 48 / Mega. BACE 4500 Training Module 1 / 1/10/2022

DNA Injection and Separation Scheme • Use 100 -200 (~150) kilo-Volt seconds (k. V-S) for injections • Typical injection on Mega. BACE is 3 k. V for 40 seconds (k. V-s) • Injection voltage should be limited to under 5 k. V • Voltages over 5 KV causes gel extrusion • 2 k. V injections give better reproducibility • Use slightly lower k. V-S for H 2 O re-suspended DNA • Salts interfere compete with DNA for injection • Air bubbles cause intermittent current dropouts • Big DNA can physically clog capillaries 49 / Mega. BACE 4500 Training Module 1 / 1/10/2022

Step 5 Run Image Module View sample run View current monitor to determine run quality 50 / Mega. BACE 4500 Training Module 1 / 1/10/2022

Routine Operation Protocols Prepare Capillaries Rehydrates the capillary arrays with water Use after installation of new arrays or after the Flush and Dry Capillaries macro Store Capillaries Stores the capillaries at 25°C with water at the cathode & anode ends Use when the Mega. BACE will not be in operation for several days 51 / Mega. BACE 4500 Training Module 1 / 1/10/2022

Routine Operation Protocols Flush & Dry Capillaries Flushes the capillaries with water and dries them with nitrogen Use when the Mega. BACE will be in long term storage Replace Capillaries High-Pressure Flush Low-Pressure Flush Focus Capillaries 52 / Mega. BACE 4500 Training Module 1 / 1/10/2022

Sequence Data Format Mega. BACE sequence files are saved in two different formats • Raw Sequence Data (Rsd) files. Upon completion of the sequencing runs, raw data (not basecalled) are saved in C: //Program Files/Amersham. Biosciences/Mega. BACE/Data • Processed or basecalled data (Esd) files. Raw data is processed by a basecaller to give the final sequence. Basecalled data is saved in C: //Program Files/Amersham. Biosciences/Mega. BACE/Analyzed. Data 53 / Mega. BACE 4500 Training Module 1 / 1/10/2022

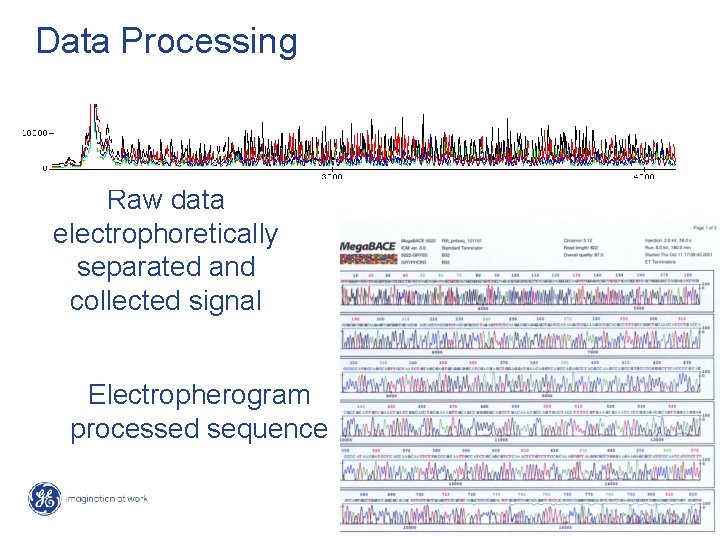
Data Processing Raw data electrophoretically separated and collected signal Electropherogram processed sequence 54 / Mega. BACE 4500 Training Module 1 / 1/10/2022

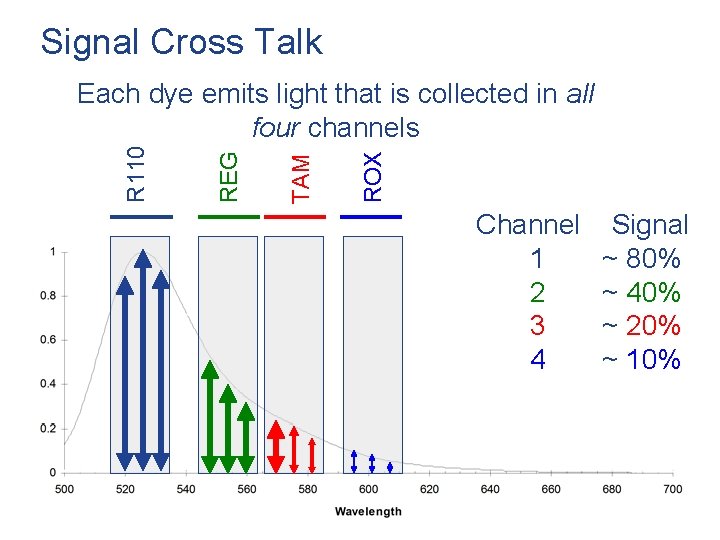
Signal Cross Talk ROX TAM REG R 110 Each dye emits light that is collected in all four channels Channel 1 2 3 4 Signal ~ 80% ~ 40% ~ 20% ~ 10% 55 / Mega. BACE 4500 Training Module 1 / 1/10/2022

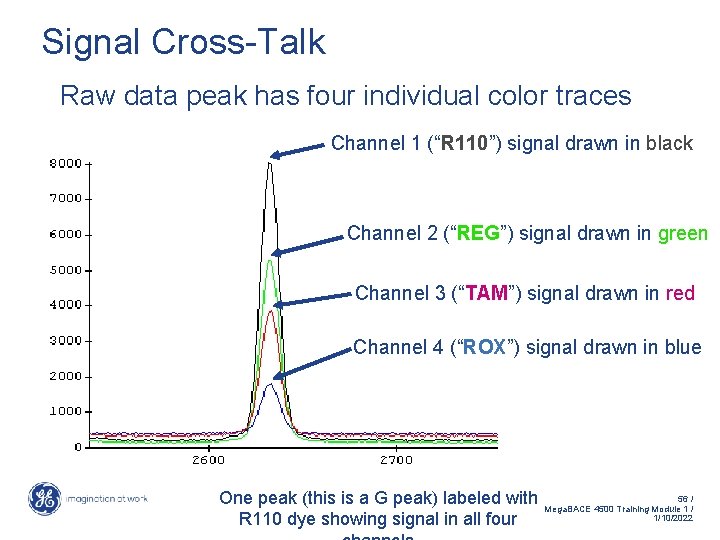
Signal Cross-Talk Raw data peak has four individual color traces Channel 1 (“R 110”) signal drawn in black Channel 2 (“REG”) signal drawn in green Channel 3 (“TAM”) signal drawn in red Channel 4 (“ROX”) signal drawn in blue One peak (this is a G peak) labeled with R 110 dye showing signal in all four 56 / Mega. BACE 4500 Training Module 1 / 1/10/2022

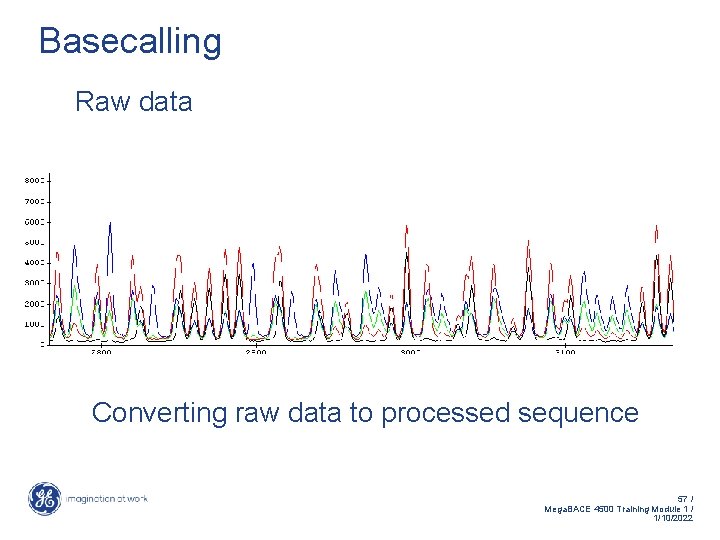
Basecalling Raw data Converting raw data to processed sequence 57 / Mega. BACE 4500 Training Module 1 / 1/10/2022

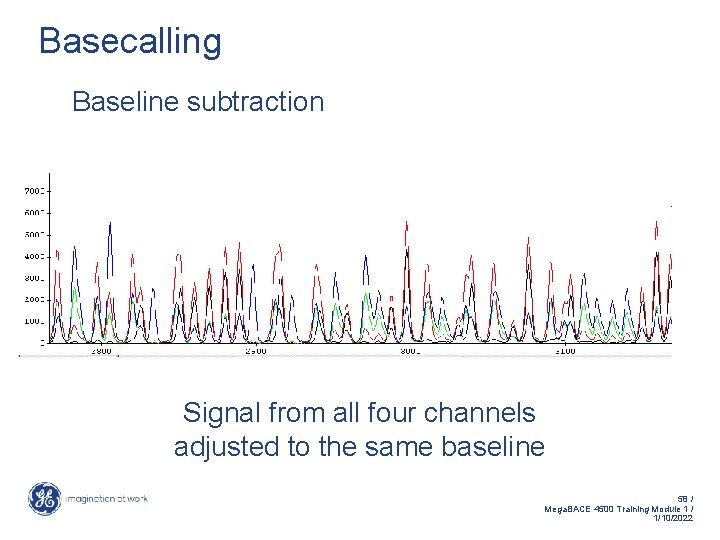
Basecalling Baseline subtraction Signal from all four channels adjusted to the same baseline 58 / Mega. BACE 4500 Training Module 1 / 1/10/2022

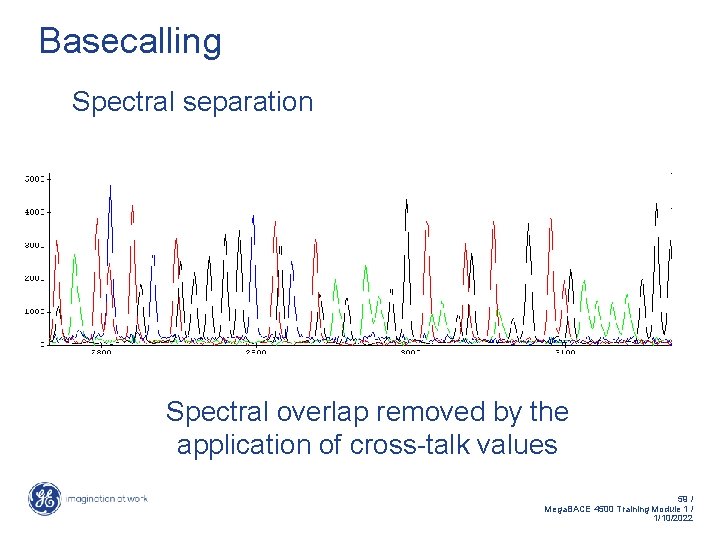
Basecalling Spectral separation Spectral overlap removed by the application of cross-talk values 59 / Mega. BACE 4500 Training Module 1 / 1/10/2022

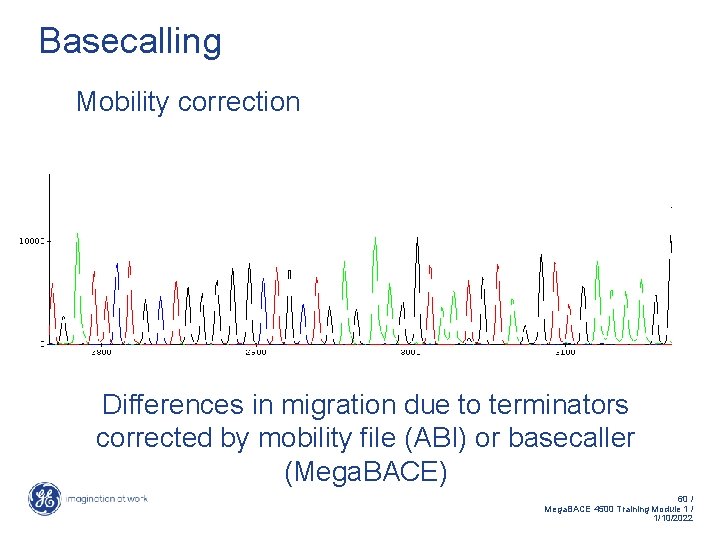
Basecalling Mobility correction Differences in migration due to terminators corrected by mobility file (ABI) or basecaller (Mega. BACE) 60 / Mega. BACE 4500 Training Module 1 / 1/10/2022

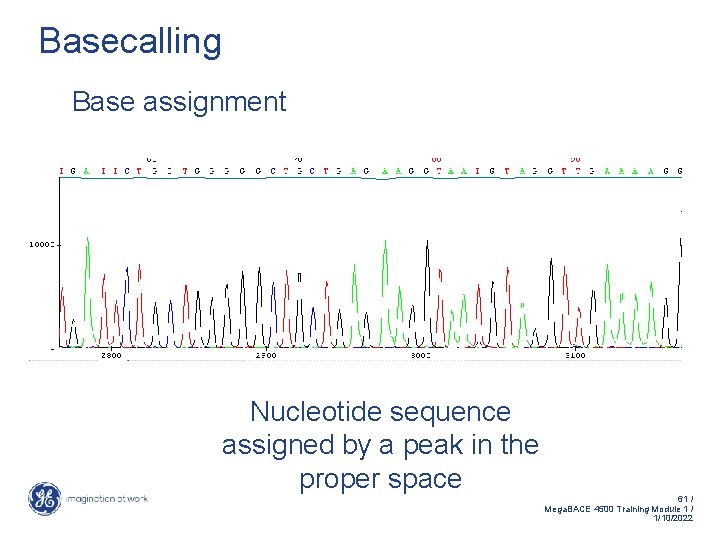
Basecalling Base assignment Nucleotide sequence assigned by a peak in the proper space 61 / Mega. BACE 4500 Training Module 1 / 1/10/2022

Mega. BACE Sequence Analyzer v 3. 0 Features • Toolbars for customizing displayed electropherogram(s) • Turn parts of the trace ON and OFF • Setting start and stop points for basecalling • Ability to view/import/export from and to other data formats • Sequence Editing • International Union of Biochemists (IUB) code display • Batch printing and print preview 62 / Mega. BACE 4500 Training Module 1 / 1/10/2022

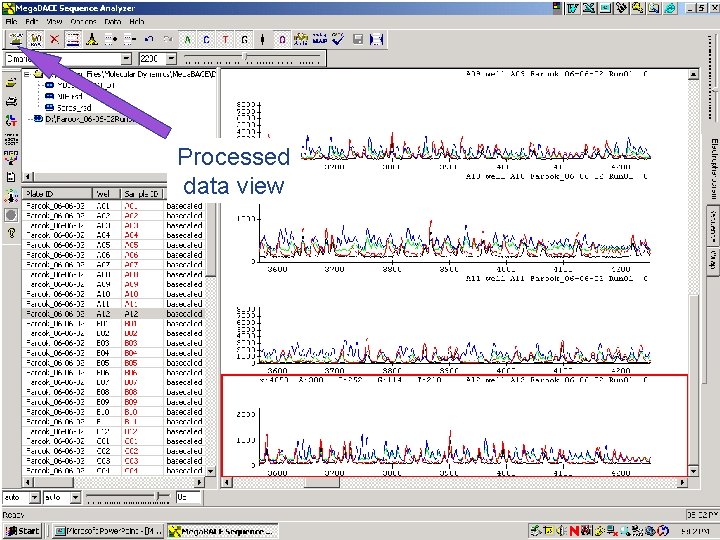
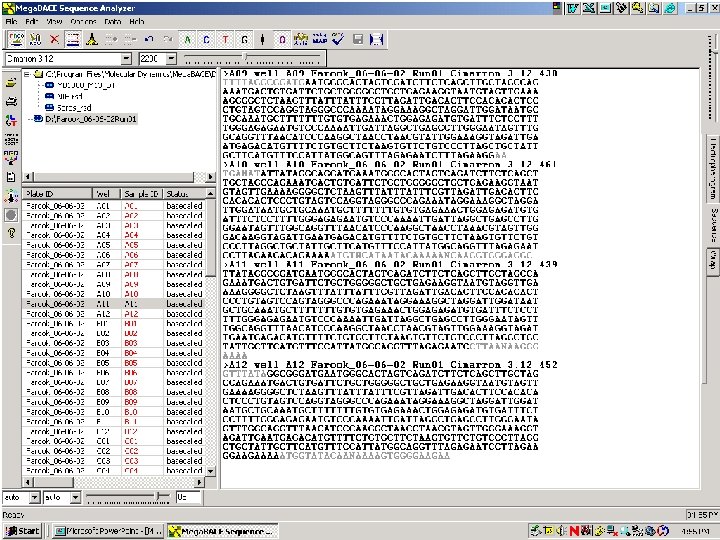
Processed data view 63 / Mega. BACE 4500 Training Module 1 / 1/10/2022

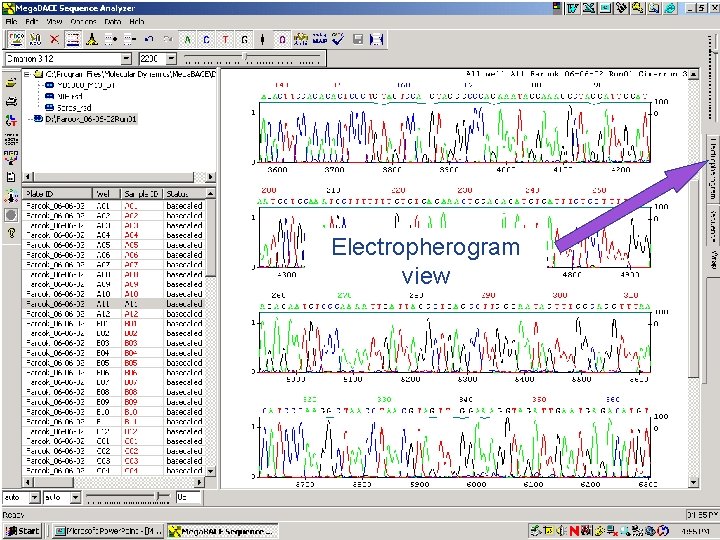
Electropherogram view 64 / Mega. BACE 4500 Training Module 1 / 1/10/2022

65 / Mega. BACE 4500 Training Module 1 / 1/10/2022

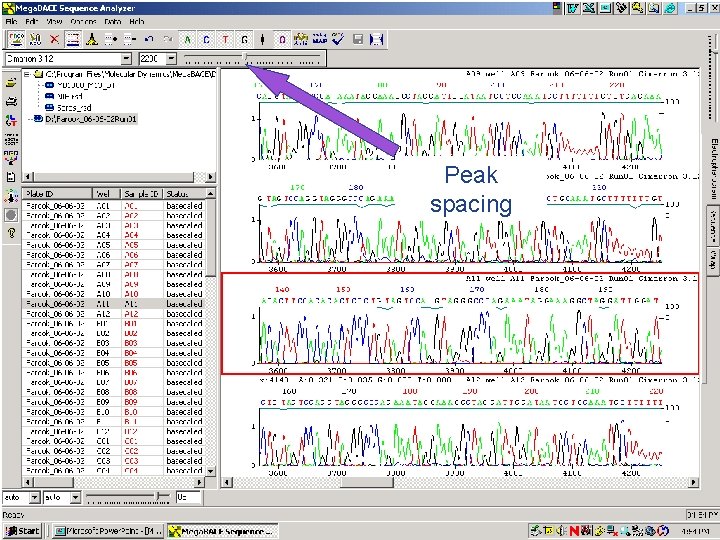
Peak spacing 66 / Mega. BACE 4500 Training Module 1 / 1/10/2022

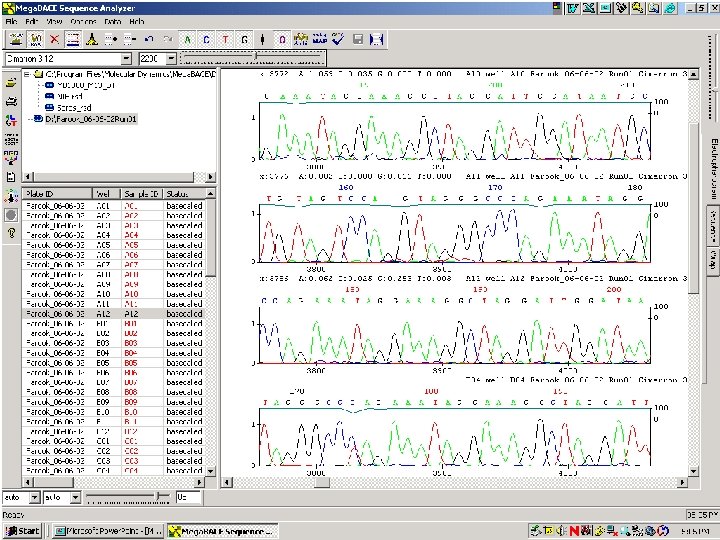
67 / Mega. BACE 4500 Training Module 1 / 1/10/2022

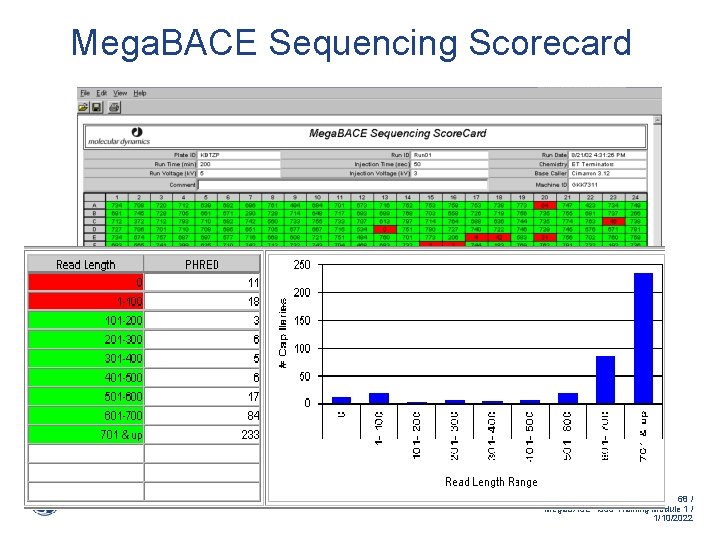
Mega. BACE Sequencing Scorecard 384 well format 68 / Mega. BACE 4500 Training Module 1 / 1/10/2022