Sequences Sequences and what they might mean Comparative




















- Slides: 41

Sequences, Sequences and what they might mean

Comparative Genomics Ø Gene functions have been conserved across evolution Ø Nature solves a problem, it rarely solves it again.

Eyeless Mutation http: //www. ucm. es/info/genetica/AVG/practicas/Drosophila. htm

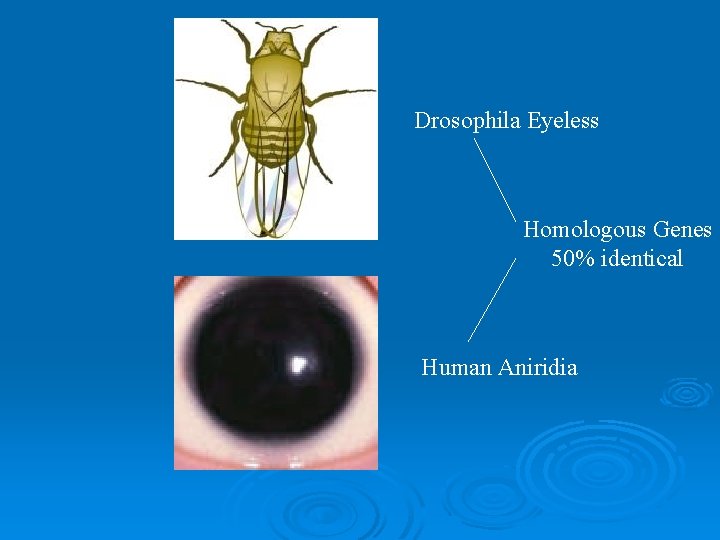
Drosophila Eyeless Homologous Genes 50% identical Human Aniridia

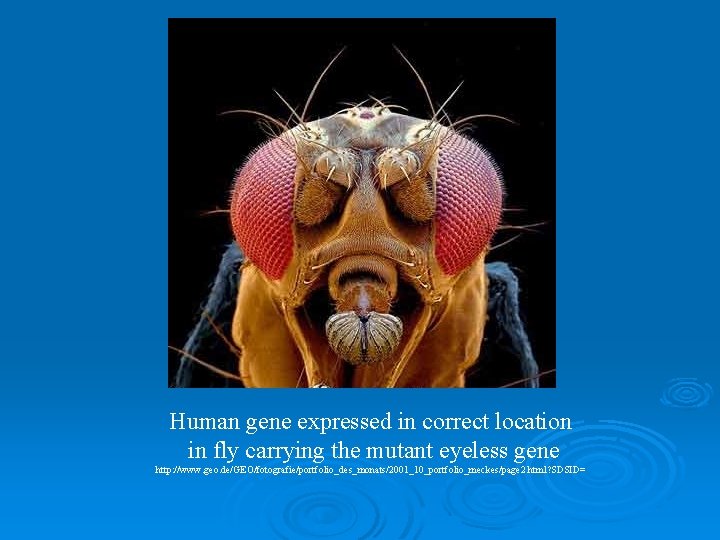
Human gene expressed in correct location in fly carrying the mutant eyeless gene http: //www. geo. de/GEO/fotografie/portfolio_des_monats/2001_10_portfolio_meckes/page 2. html? SDSID=

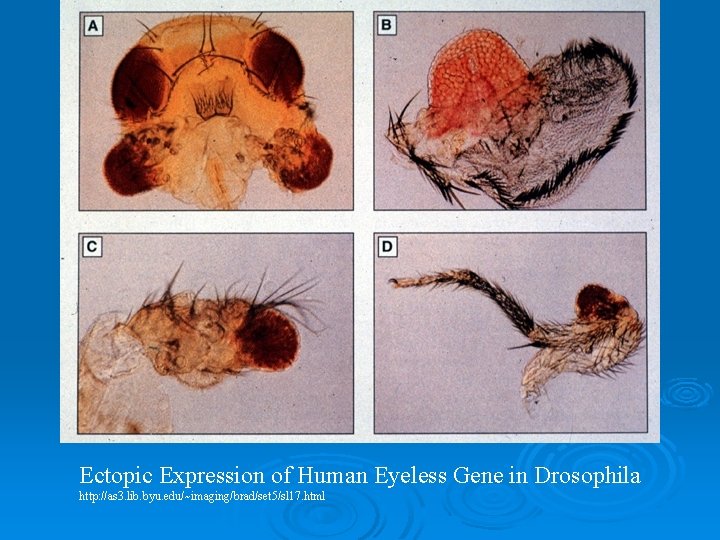
Ectopic Expression of Human Eyeless Gene in Drosophila http: //as 3. lib. byu. edu/~imaging/brad/set 5/sl 17. html


Flies with human eyeless homolog do not develop “human” eyes

Develop compound eyes, even with human homolog driving development

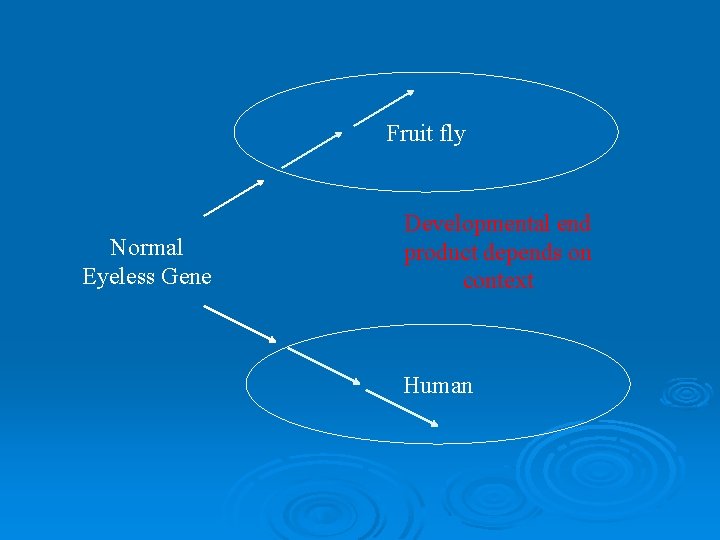
Fruit fly Normal Eyeless Gene Developmental end product depends on context Human

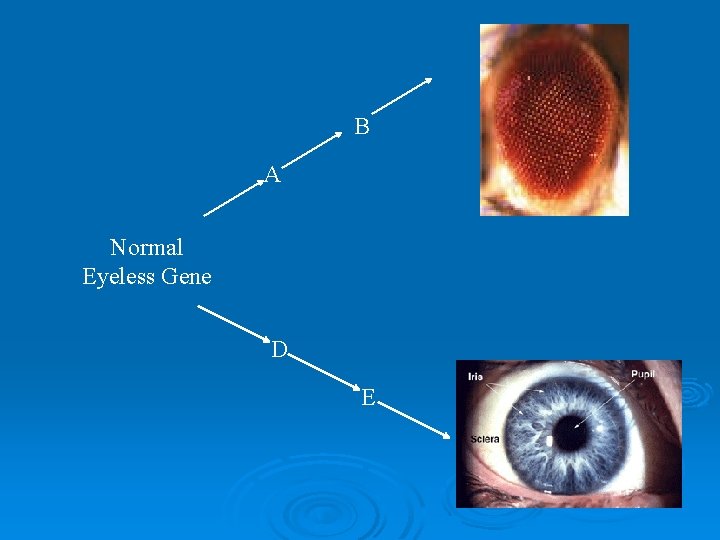
B A Normal Eyeless Gene D E

Evolution & Genes Ø The function of many genes is conserved across immense evolutionary distances. Ø The function of a gene is affected by its environment. Ø Many gene products function as part of a cascade or pathway.

“Nothing in Biology Makes Sense Except in the Light of Evolution” (Theodosius Dobzhansky)

Map Genes in the Context of Chromosomes

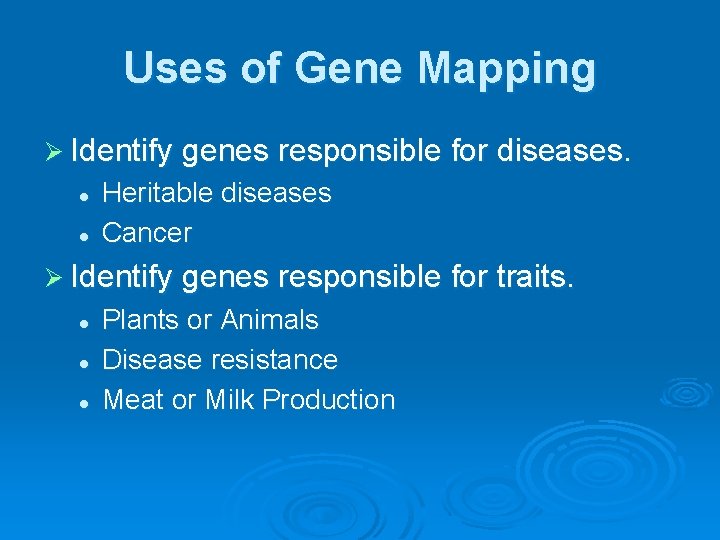
Uses of Gene Mapping Ø Identify genes responsible for diseases. l l Heritable diseases Cancer Ø Identify genes responsible for traits. l l l Plants or Animals Disease resistance Meat or Milk Production

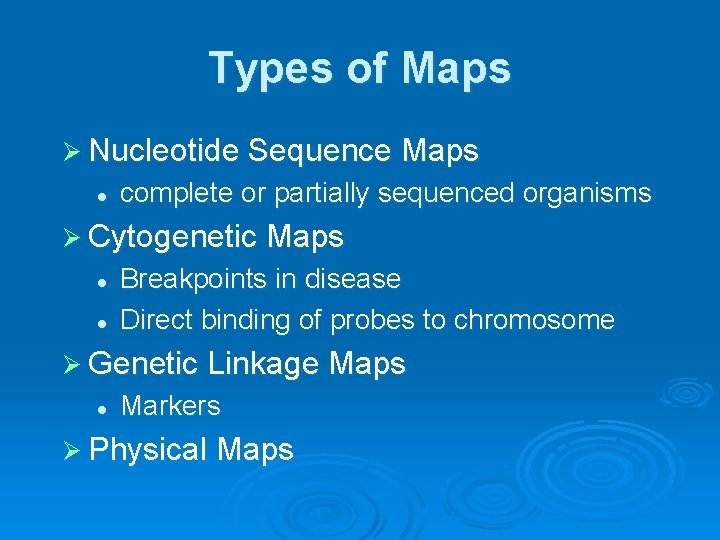
Types of Maps Ø Nucleotide Sequence Maps l complete or partially sequenced organisms Ø Cytogenetic Maps l l Breakpoints in disease Direct binding of probes to chromosome Ø Genetic Linkage Maps l Markers Ø Physical Maps

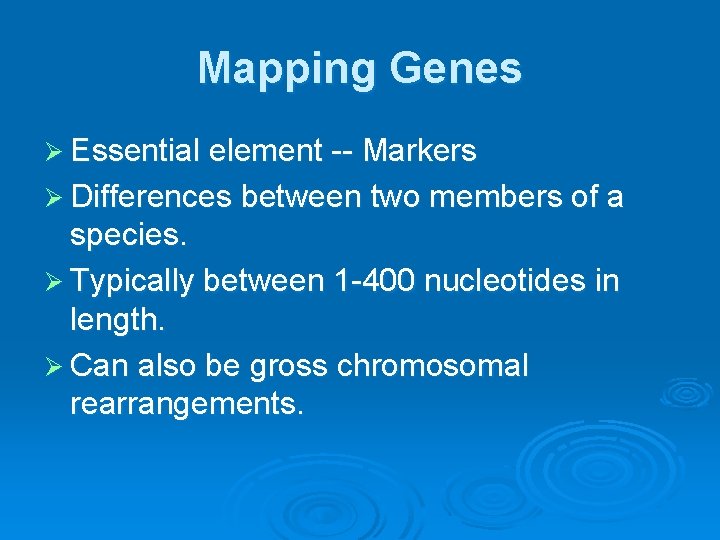
Mapping Genes Ø Essential element -- Markers Ø Differences between two members of a species. Ø Typically between 1 -400 nucleotides in length. Ø Can also be gross chromosomal rearrangements.

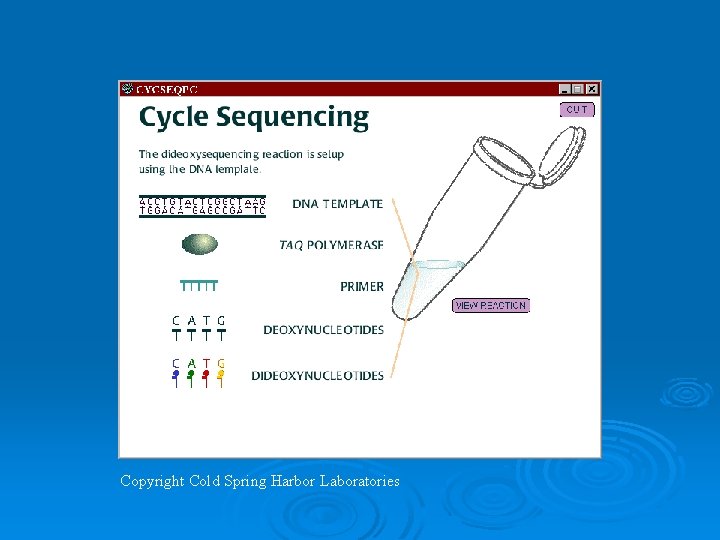
DNA Sequencing Ø Two methods originally developed: l l l Chemical or Maxim and Gilbert method. Dideoxynucleotide or Sanger method. Both Gilber and Sanger won Nobel prizes for developing these techniques. Ø Dideoxynucleotide method is by far the most common employed today.

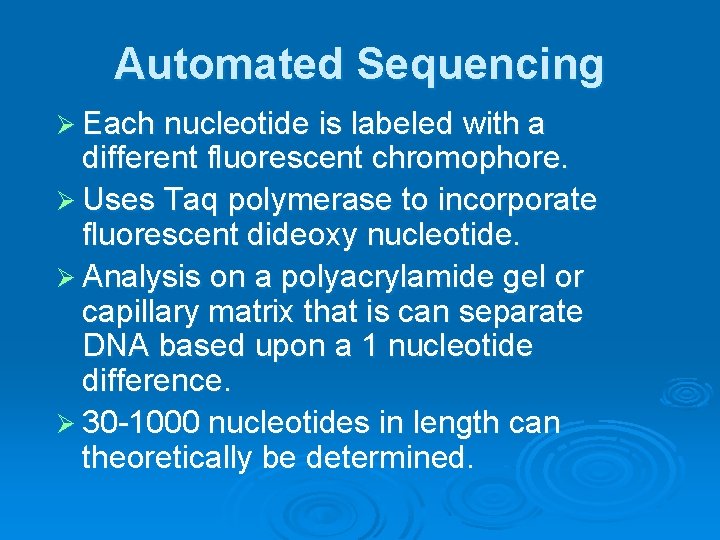
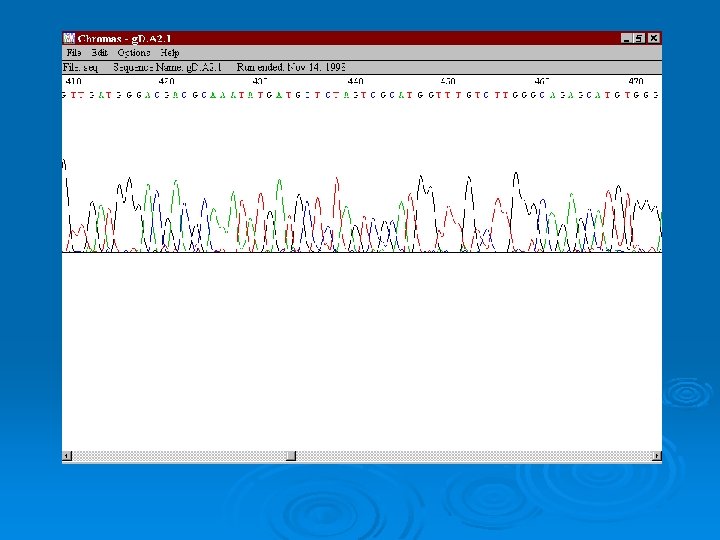
Automated Sequencing Ø Each nucleotide is labeled with a different fluorescent chromophore. Ø Uses Taq polymerase to incorporate fluorescent dideoxy nucleotide. Ø Analysis on a polyacrylamide gel or capillary matrix that is can separate DNA based upon a 1 nucleotide difference. Ø 30 -1000 nucleotides in length can theoretically be determined.

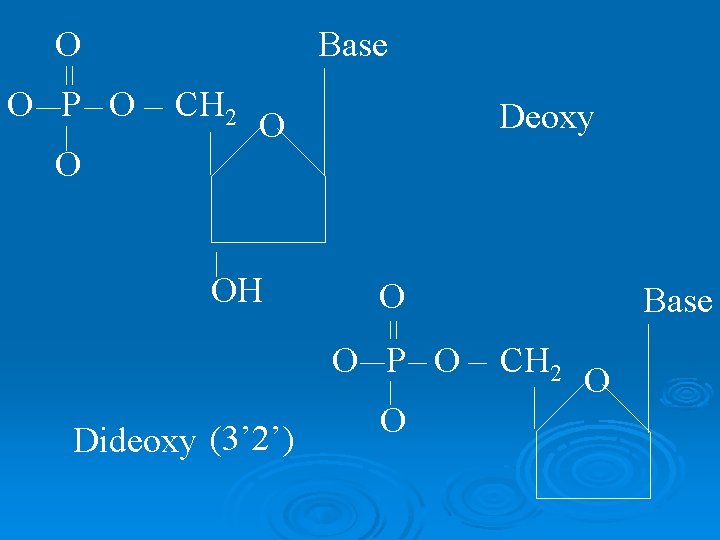
O O P O O Base CH 2 Deoxy O OH O O P O Dideoxy (3’ 2’) O Base CH 2 O

Copyright Cold Spring Harbor Laboratories

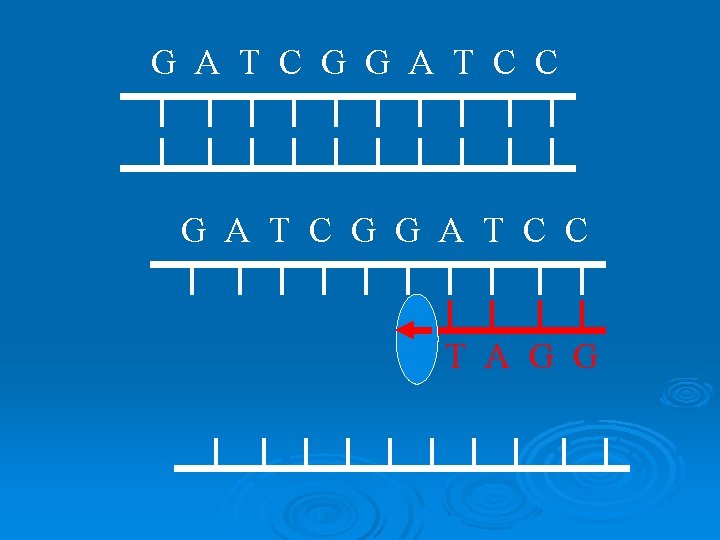
G A T C G G A T C C T A G G

G A T C G G A T C C C C T A G G dd

G A T C G G A T C C GC C T A G G dd


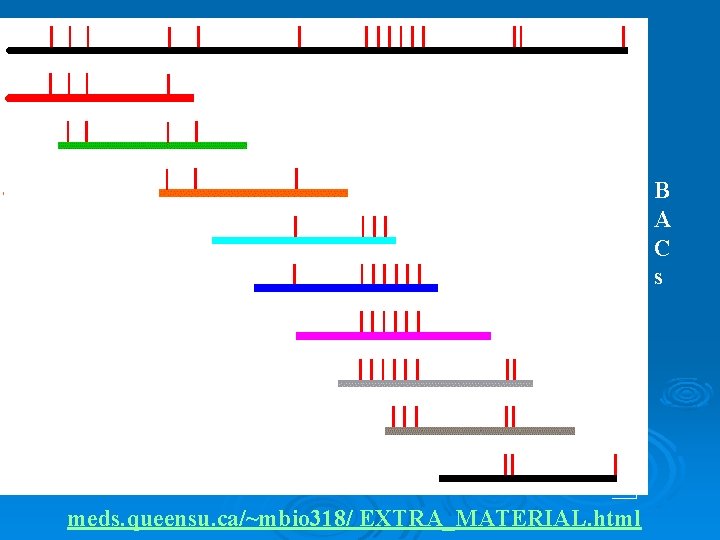
Nucleotide Sequencing Ø Complete Genome Sequencing l Bacterial Artificial Chromosomes (BAC) • Can grow in bacteria • Have large inserts 100, 000 -300, 000 nucleotides l l Collect a large library of BACs Restriction Endonuclease (RE) Map • RE - cuts DNA at specific sites • Eco. RI cuts at GAATTC

B A C s meds. queensu. ca/~mbio 318/ EXTRA_MATERIAL. html

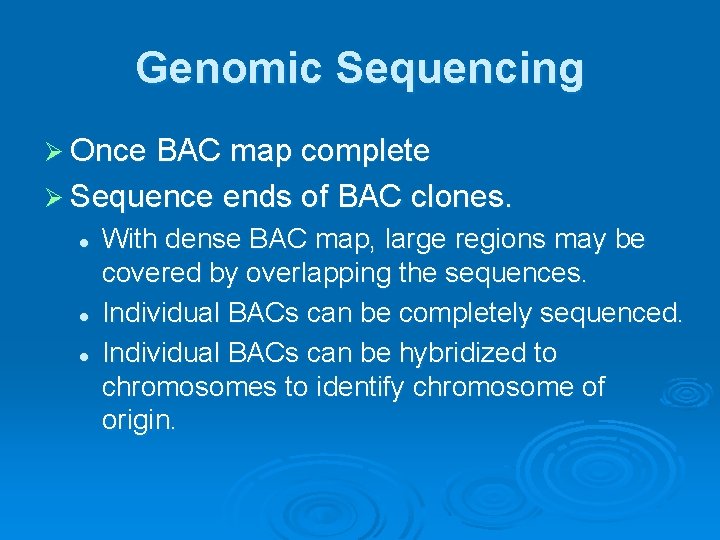
Genomic Sequencing Ø Once BAC map complete Ø Sequence ends of BAC clones. l l l With dense BAC map, large regions may be covered by overlapping the sequences. Individual BACs can be completely sequenced. Individual BACs can be hybridized to chromosomes to identify chromosome of origin.

Recombination Ø Permits Mapping by providing: l l Linkage groups Distances Ø During Meiosis

Recombination Ø Linkage Groups - markers that tend to remain together. Ø Distance - the further apart two markers lie, the more often recombination will occur between those markers. Ø Markers on the same chromosome can be so far apart that they appear in different linkage groups.

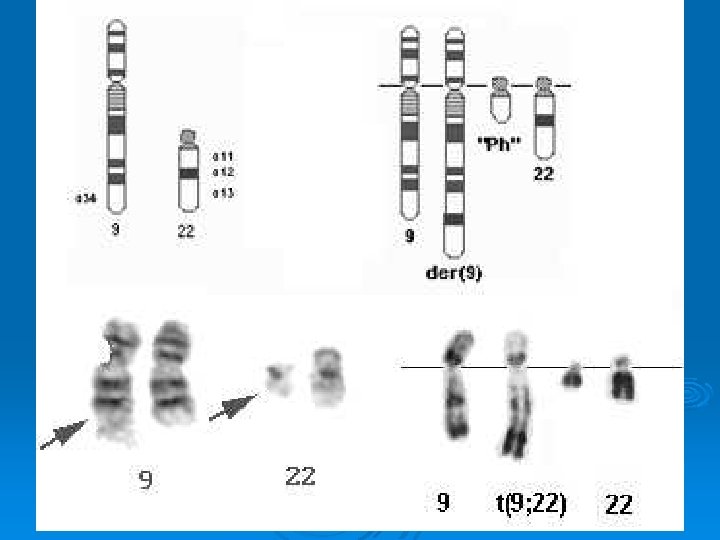
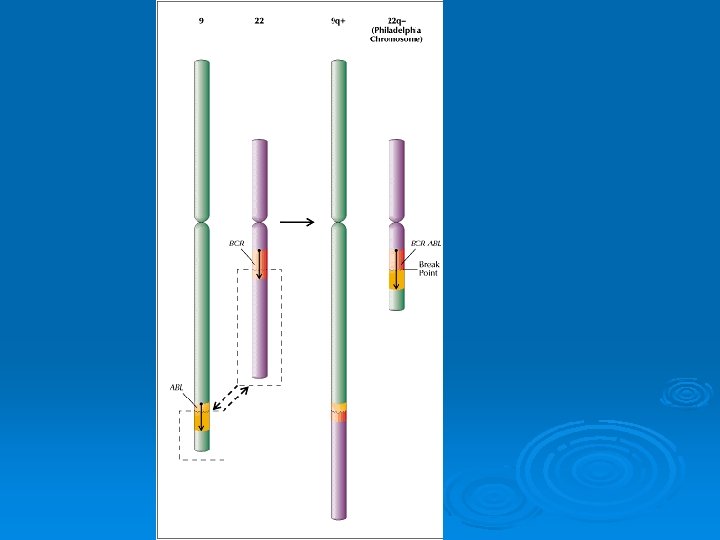
Cytogenetic Mapping Philadelphia Chromosome Ø Chronic myelogenous leukemia (CML) is characterized by a reciprocal translocation between chromosomes 9 and 22 that produces the Philadelphia chromosome. Invariably there is disease progression, with loss of the capacity for terminal differentiation by the hematopoietic stem cell, resulting in an acute leukemia. Ø www. clubstewart. co. uk/cml. htm



Genetic Linkage Maps Polymorphism Ø Polymorphisms l l Polymorphism - an allele present in a population that exhibits multiple forms. Monomorph - Single form in a population. Common. Typically result in no effect on survival of individual.

Types of Markers Ø Single Nucleotide Polymorphisms (SNPs) l Occur approximately every 100 -300 bp in humans. Names: rs 8111765

Markers Ø Microsatellites or Tandem Repeats l l acg. CACACAtgc acg. CACAtgc

Physical Maps Ø Sequence Tagged Sites (STSs) l l Use Polymerase Chain Reaction (PCR). Primers based upon: • random sequence • expressed sequence tag l l l Amplify from library of clones containing large inserts (BAC). Relate to BAC map. If more than one on same clone, then close together

Types of questions Ø Where does gene X exist within the genome of organism Y? What are some flanking markers? Ø Which genes exist on a chromosome, and in what order do they appear? Ø Show the genes that exist in region R of the chromosome. Show me the corresponding sequence data for that region. Ø Display the region of a chromosome between points A and B. Ø Show both the cytogenetic and sequence map for that region, aligned to each other based on markers that have been placed on both maps. Ø What is the distance between two genes? (Note: scale depends on the type of map on which those genes have been placed. ) Ø I know the cytogenetic location of gene X. What is the corresponding physical location? http: //www. ncbi. nlm. nih. gov/mapview/static/Map. Viewer. Help. html

Genes and Maps Ø Bcl 2 Ø Map Link Ø OMIM Link Ø Syntenic Relationships - tendency of closely linked genes to remain linked during evolution.

GO evidence codes

Pathways KEGG pathways Examine Cell Adhesion Molecules -What does JAM 3 appear to interact with? -What information is available if you click on JAM 2? What motifs are found in JAM 2? Click on Tight Junction -What molecules appear to interact with the JAMJAM dimer? What is ZO-1 Ø