Sequence formats Say we get this protein sequence









- Slides: 18

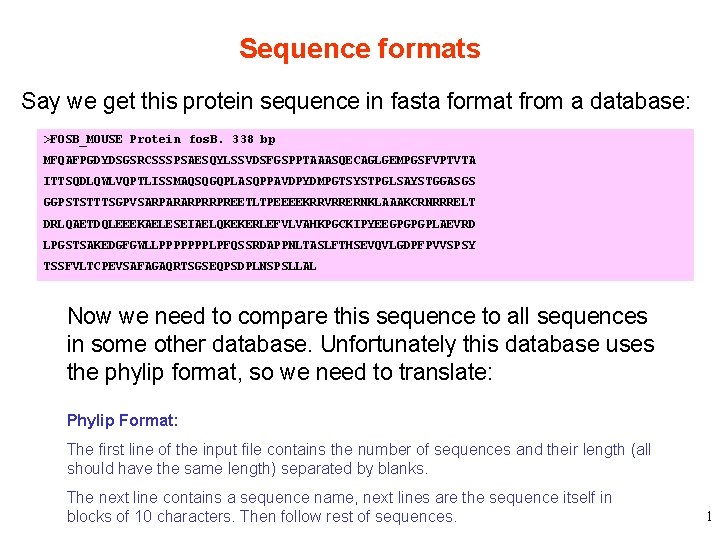
Sequence formats Say we get this protein sequence in fasta format from a database: >FOSB_MOUSE Protein fos. B. 338 bp MFQAFPGDYDSGSRCSSSPSAESQYLSSVDSFGSPPTAAASQECAGLGEMPGSFVPTVTA ITTSQDLQWLVQPTLISSMAQSQGQPLASQPPAVDPYDMPGTSYSTPGLSAYSTGGASGS GGPSTSTTTSGPVSARPARARPRRPREETLTPEEEEKRRVRRERNKLAAAKCRNRRRELT DRLQAETDQLEEEKAELESEIAELQKEKERLEFVLVAHKPGCKIPYEEGPGPGPLAEVRD LPGSTSAKEDGFGWLLPPPPPPPLPFQSSRDAPPNLTASLFTHSEVQVLGDPFPVVSPSY TSSFVLTCPEVSAFAGAQRTSGSEQPSDPLNSPSLLAL Now we need to compare this sequence to all sequences in some other database. Unfortunately this database uses the phylip format, so we need to translate: Phylip Format: The first line of the input file contains the number of sequences and their length (all should have the same length) separated by blanks. The next line contains a sequence name, next lines are the sequence itself in blocks of 10 characters. Then follow rest of sequences. 1

Sequence formats >FOSB_MOUSE Protein fos. B. 338 bp MFQAFPGDYDSGSRCSSSPSAESQYLSSVDSFGSPPTAAASQECAGLGEMPGSFVPTVTA ITTSQDLQWLVQPTLISSMAQSQGQPLASQPPAVDPYDMPGTSYSTPGLSAYSTGGASGS GGPSTSTTTSGPVSARPARARPRRPREETLTPEEEEKRRVRRERNKLAAAKCRNRRRELT DRLQAETDQLEEEKAELESEIAELQKEKERLEFVLVAHKPGCKIPYEEGPGPGPLAEVRD Fasta LPGSTSAKEDGFGWLLPPPPPPPLPFQSSRDAPPNLTASLFTHSEVQVLGDPFPVVSPSY TSSFVLTCPEVSAFAGAQRTSGSEQPSDPLNSPSLLAL So we copy and paste and reformat the sequence: 1 338 FOSB_MOUSE MFQAFPGDYD SGSRCSSSPS AESQYLSSVD SFGSPPTAAA SQECAGLGEM PGSFVPTVTA ITTSQDLQWL VQPTLISSMA QSQGQPLASQ PPAVDPYDMP Phylip GTSYSTPGLS AYSTGGASGS GGPSTSTTTS GPVSARPARA RPRRPREETL TPEEEEKRRV RRERNKLAAA KCRNRRRELT DRLQAETDQL EEEKAELESE IAELQKEKER LEFVLVAHKP GCKIPYEEGP GPGPLAEVRD LPGSTSAKED GFGWLLPPPP PPPLPFQSSR DAPPNLTASL FTHSEVQVLG DPFPVVSPSY TSSFVLTCPE VSAFAGAQRT SGSEQPSDPL NSPSLLAL and all is well. Then our boss says “Do it for these 5000 sequences. ” 2

We need automatic filter! • A program that reads any number of fasta sequences and converts them into phylip format (want to run sequences through a filter) • Program structure: 1. Open fasta file 2. Parse file to extract needed information 3. Create and save phylip file • We will use this definition for the fasta format: – The header starts with > – The word immediately following the ">" is a unique ID; next two words are the species name, the rest of the header is a description. – All lines of text are shorter than 80 characters. 3

Pseudo-code fasta→phylip filter 1. Open and parse fasta file 2. From each header extract sequence ID and name 1. Open phylip file 2. Write “ 1” followed by sequence length 3. Write sequence ID 4. Write sequence in blocks of 10 5. Close file 4

The other way too: pseudo-code phylip→fasta filter 1. Open phylip file 2. Find first non-empty line, ignore! 3. Parse next line and extract first word (sequence ID) 1. Read rest of line and following lines to get the sequence, skipping blanks 2. Read next sequences 4. Open fasta file, and for each sequence: 1. Write “>” followed by sequence name 2. Write sequence in lines of 80 5. Close files 5

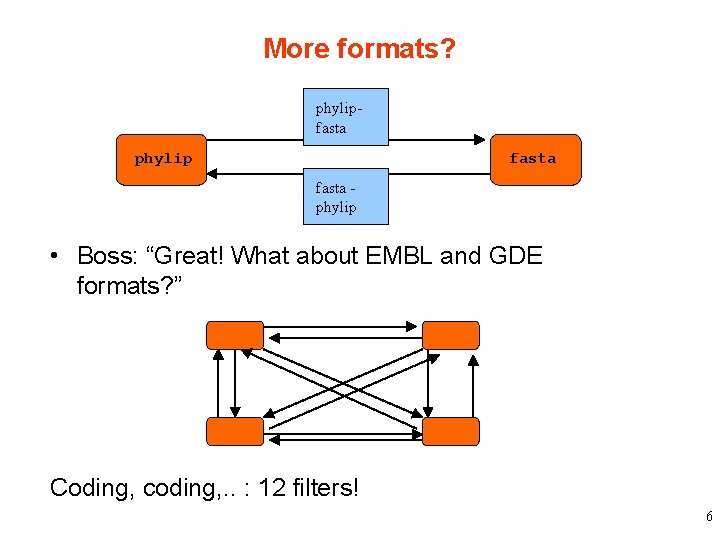
More formats? phylipfasta phylip • Boss: “Great! What about EMBL and GDE formats? ” Coding, coding, . . : 12 filters! 6

Still more formats? • Boss: “Super. And Genebank and Clustal. W. . ? ” Coding, coding, . . : 30 filters • Next new format = 12 new filters! • This doesn’t scale. 7

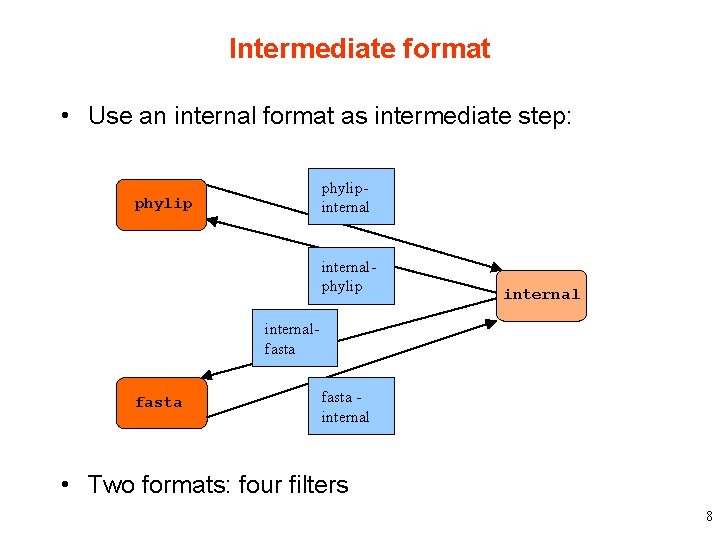
Intermediate format • Use an internal format as intermediate step: phylipinternal phylip internalfasta internal • Two formats: four filters 8

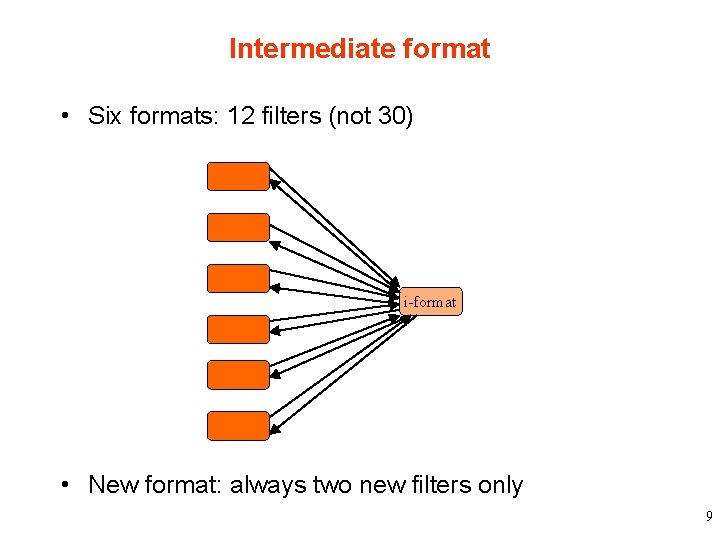
Intermediate format • Six formats: 12 filters (not 30) i-format • New format: always two new filters only 9

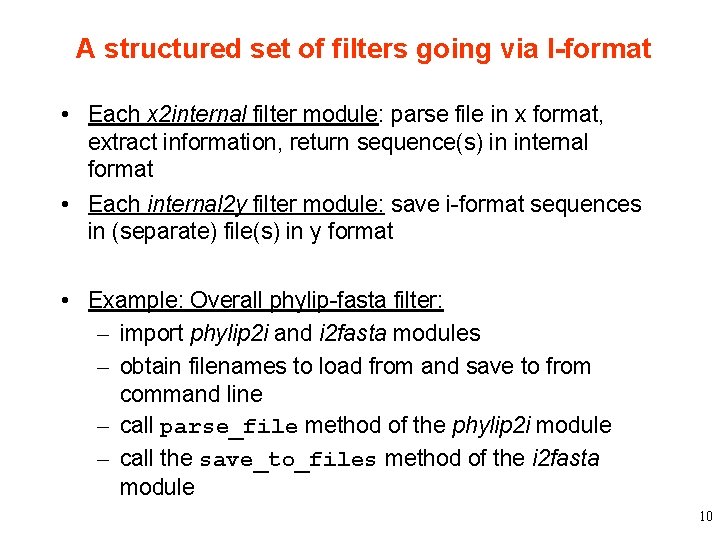
A structured set of filters going via I-format • Each x 2 internal filter module: parse file in x format, extract information, return sequence(s) in internal format • Each internal 2 y filter module: save i-format sequences in (separate) file(s) in y format • Example: Overall phylip-fasta filter: – import phylip 2 i and i 2 fasta modules – obtain filenames to load from and save to from command line – call parse_file method of the phylip 2 i module – call the save_to_files method of the i 2 fasta module 10

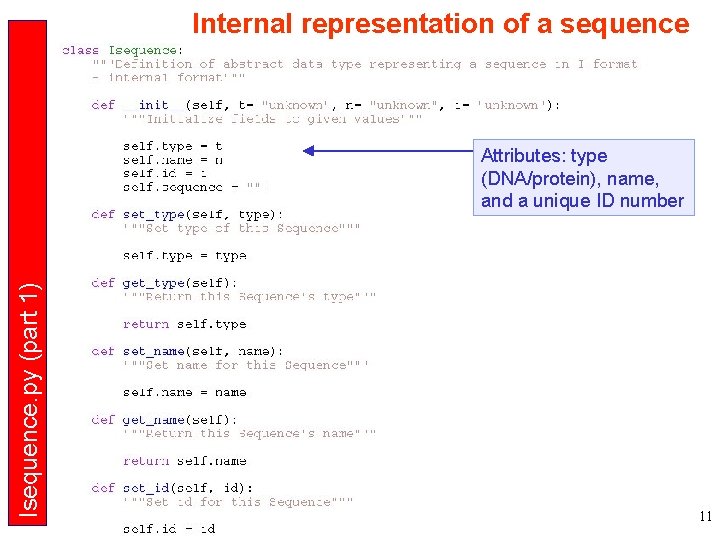
Internal representation of a sequence Isequence. py (part 1) Attributes: type (DNA/protein), name, and a unique ID number 11

Isequence. py (part 2) 12

Example: fasta/phylip filter fasta 2 i. py First fasta 2 internal. Each x 2 internal filter module: parse file in x format, extract information, return sequence(s) in internal format 13

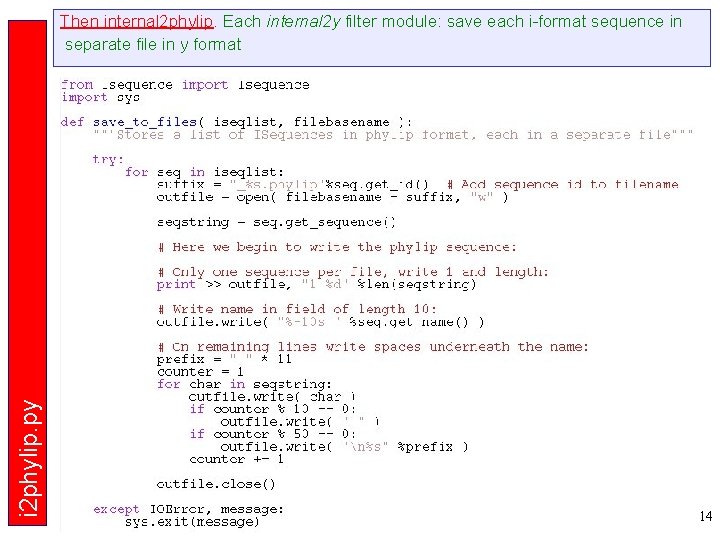
i 2 phylip. py Then internal 2 phylip. Each internal 2 y filter module: save each i-format sequence in separate file in y format 14

fasta 2 phylip. py Putting the parts together: Fasta/phylip filter 1. Import parse_file method from fasta 2 i module 2. Import save_to_files method from i 2 phylip module 3. Obtain filenames to load from and save to from command line 4. Call parse_file method 5. Call the save_to_files method NB: nothing in code about phylip and fasta below this point. . 15

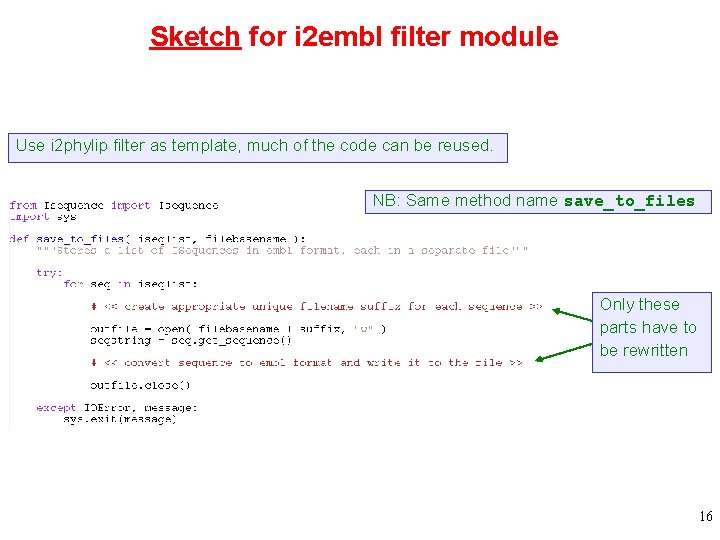
Sketch for i 2 embl filter module Use i 2 phylip filter as template, much of the code can be reused. NB: Same method name save_to_files Only these parts have to be rewritten 16

Complete fasta/embl filter (assuming we have the i 2 embl filter. . ) fasta 2 embl. py Almost the same code as the fasta 2 phylip filter: only change is that the method save_to_files is imported from new module 17

. . on to the exercises 18