Sequence Analysis with Artemis Artemis Comparison Tool ACT





- Slides: 37

Sequence Analysis with Artemis & Artemis Comparison Tool (ACT) South East Asian Training Course on Bioinformatics Applied to Tropical Diseases - 2005 (Sponsored by UNDP/World Bank/WHO/TDR) International Centre For Genetic Engineering And Biotechnology , New Delhi, INDIA

Workshop Overview of the genome sequencing and sequence analysis. Demonstration of Artemis. Hands on guided exercise in Artemis. Demonstration of ACT. Hands on guided exercise in ACT Generating ACT comparison files

The Wellcome Trust Sanger Institute • Funded by The Wellcome Trust, a registered charity. • Established in 1993 to begin the Human genome project. • First Draft (2000) complete (2003 -4) Wellcome Trust Photo Library Data release policy: All sequence data is released immediately and is freely available via the internet in order to maximise its benefit for research. http: //www. sanger. ac. uk ftp: //ftp. sanger. ac. uk/ Wellcome Trust Photo Library

Generating the complete genome sequence

Infrastructure

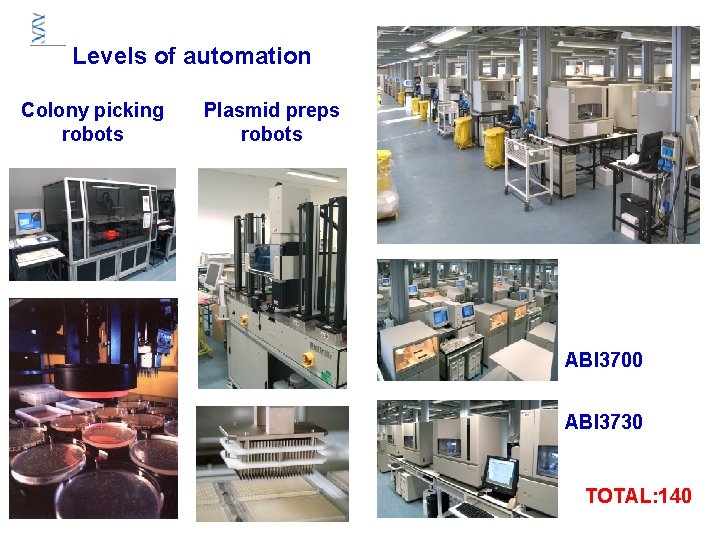
Levels of automation Colony picking robots Plasmid preps robots ABI 3700 ABI 3730 TOTAL: 140

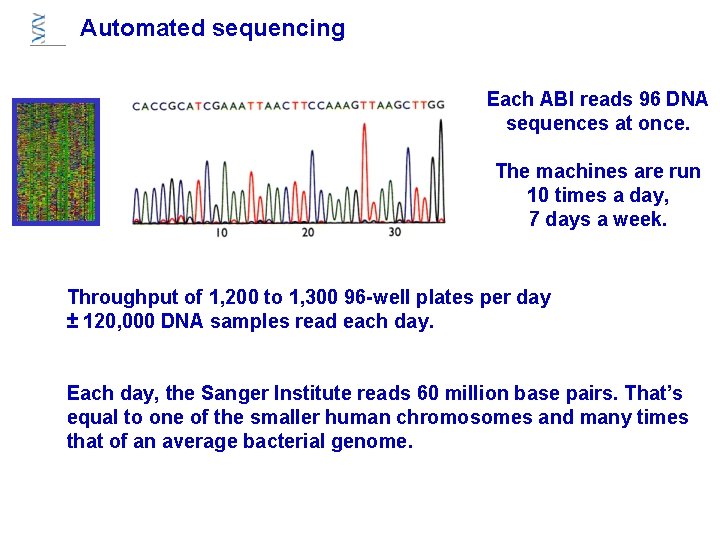
Automated sequencing Each ABI reads 96 DNA sequences at once. The machines are run 10 times a day, 7 days a week. Throughput of 1, 200 to 1, 300 96 -well plates per day ± 120, 000 DNA samples read each day. Each day, the Sanger Institute reads 60 million base pairs. That’s equal to one of the smaller human chromosomes and many times that of an average bacterial genome.

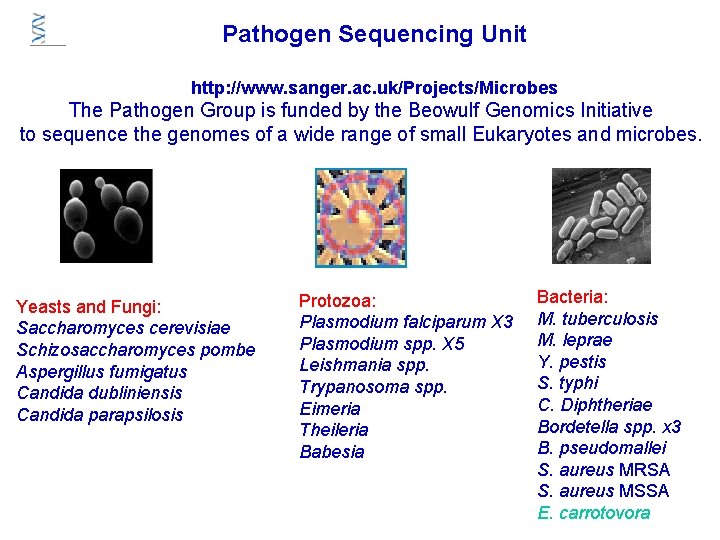
Pathogen Sequencing Unit http: //www. sanger. ac. uk/Projects/Microbes The Pathogen Group is funded by the Beowulf Genomics Initiative to sequence the genomes of a wide range of small Eukaryotes and microbes. Yeasts and Fungi: Saccharomyces cerevisiae Schizosaccharomyces pombe Aspergillus fumigatus Candida dubliniensis Candida parapsilosis Protozoa: Plasmodium falciparum X 3 Plasmodium spp. X 5 Leishmania spp. Trypanosoma spp. Eimeria Theileria Babesia Bacteria: M. tuberculosis M. leprae Y. pestis S. typhi C. Diphtheriae Bordetella spp. x 3 B. pseudomallei S. aureus MRSA S. aureus MSSA E. carrotovora

Sequencing strategy and assembly

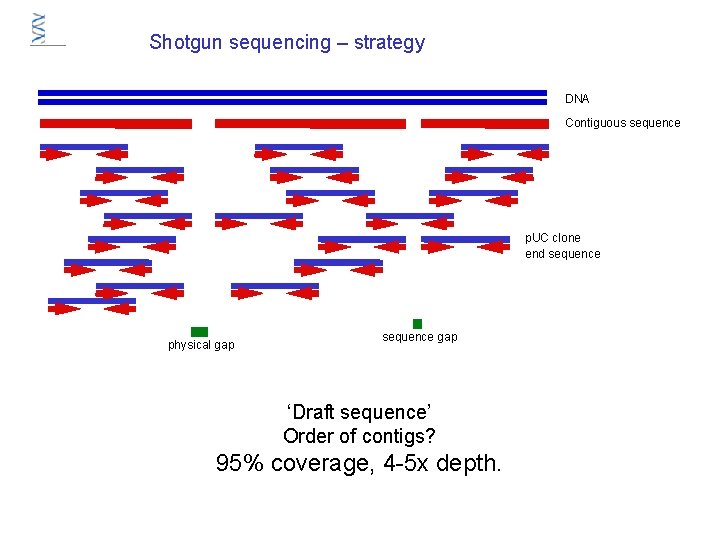
Shotgun sequencing – strategy DNA Contiguous sequence p. UC clone end sequence physical gap sequence gap ‘Draft sequence’ Order of contigs? 95% coverage, 4 -5 x depth.

‘A genome in a day’ ‘ 15 in a month’ ‘High-quality draft sequence’

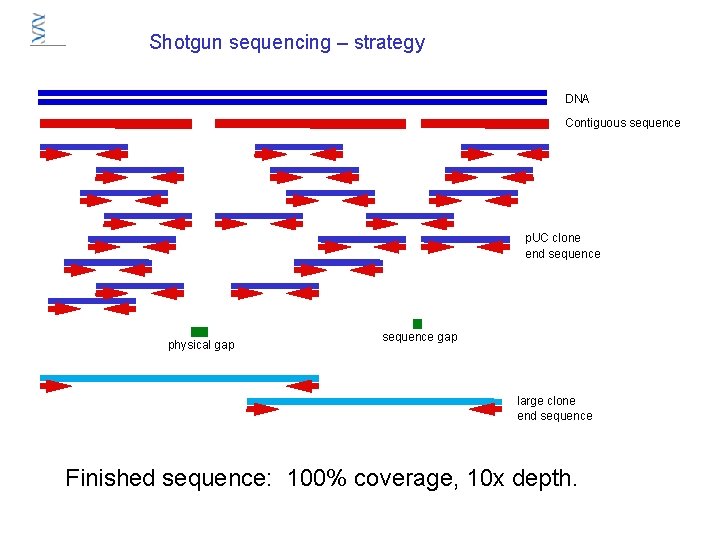
Shotgun sequencing – strategy DNA Contiguous sequence p. UC clone end sequence physical gap sequence gap large clone end sequence Finished sequence: 100% coverage, 10 x depth.

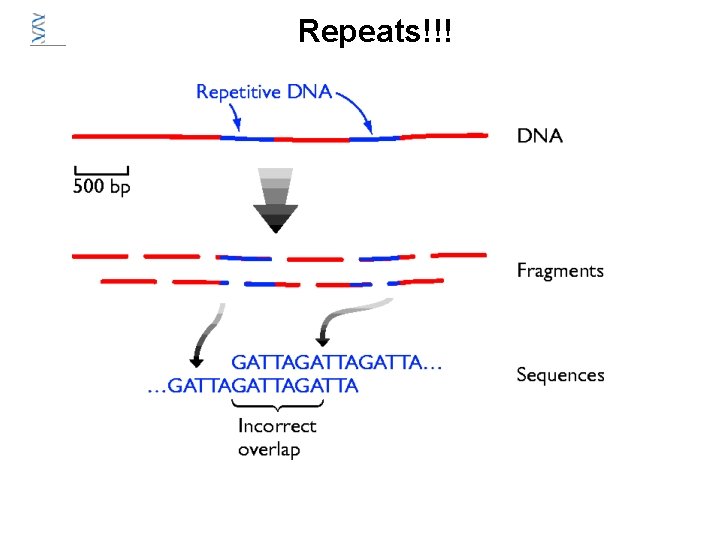
Repeats!!!

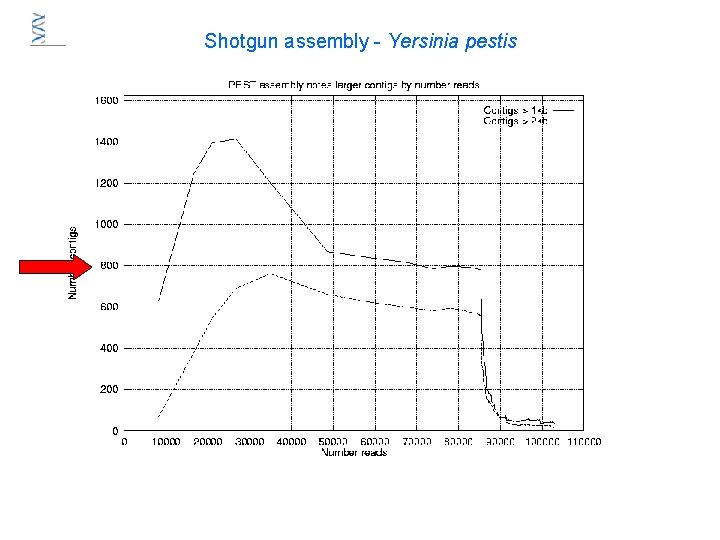
Shotgun assembly - Yersinia pestis

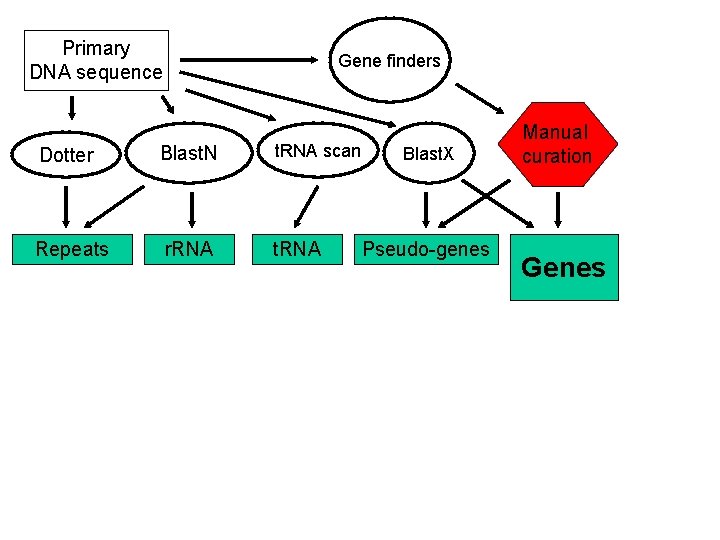
Primary DNA sequence Gene finders Dotter Blast. N t. RNA scan Repeats r. RNA t. RNA Blast. X Pseudo-genes Manual curation Genes

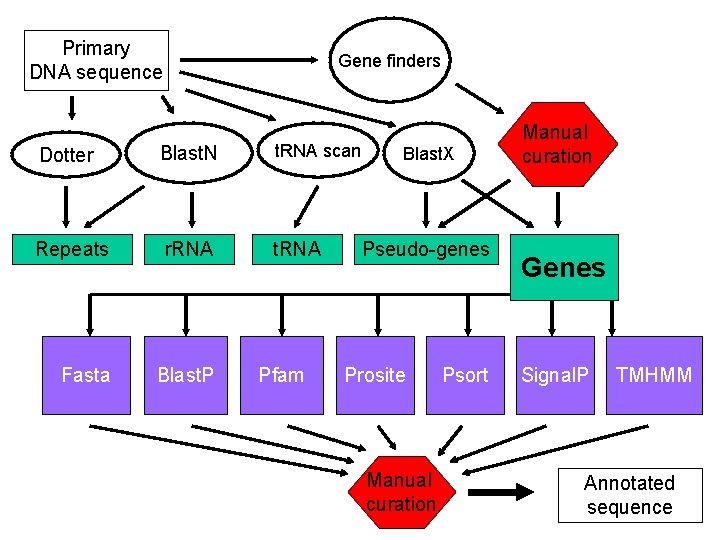
Primary DNA sequence Gene finders Dotter Blast. N t. RNA scan Repeats r. RNA t. RNA Fasta Blast. P Pfam Blast. X Pseudo-genes Prosite Manual curation Psort Manual curation Genes Signal. P TMHMM Annotated sequence

PSU Projects Organism Database entry Finished genome Annotated genome Artemis

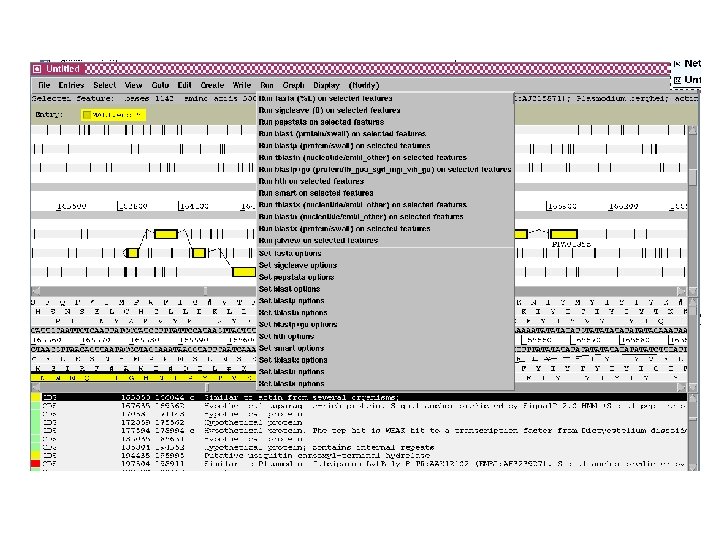
Artemis • Sequence viewer and analysis tool – Visualization of sequence features • DNA • Six frame translation – Perform and view analysis • Basic analysis • Launch more complex analysis and searches • Import and view the results of other searches

Outline of Artemis demonstration • • Artemis window features Open a genome sequence Changing the view Getting around – Goto Menu – Navigator – Feature Selector • Basic analysis – Edit a feature – Fasta search – Show feature plots

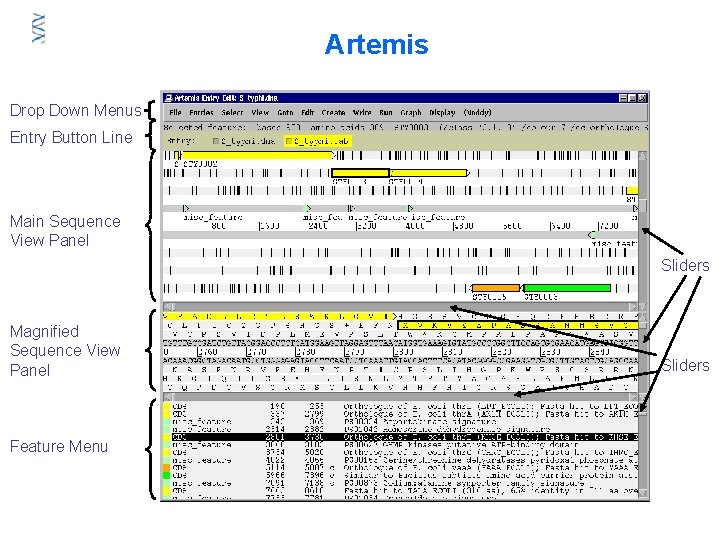
Artemis Drop Down Menus Entry Button Line Main Sequence View Panel Sliders Magnified Sequence View Panel Feature Menu Sliders

Artemis

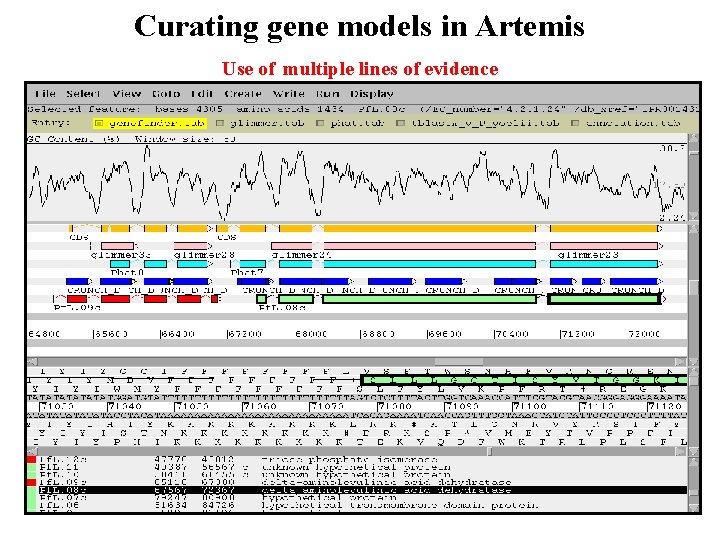
Curating gene models in Artemis Use of multiple lines of evidence

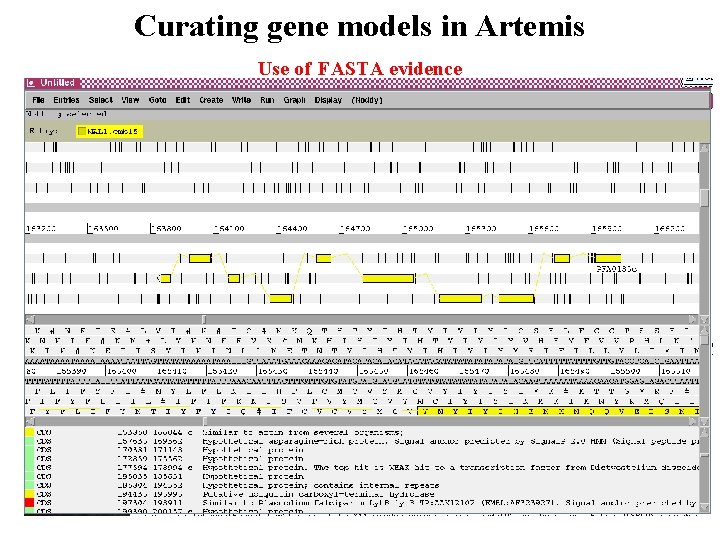
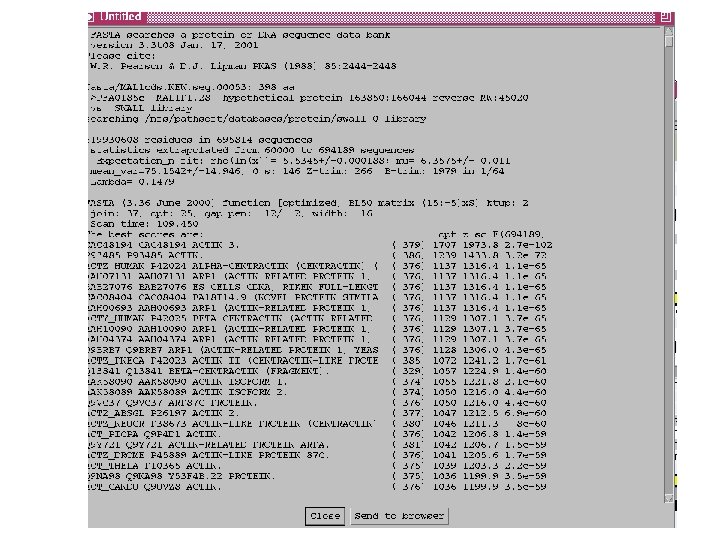
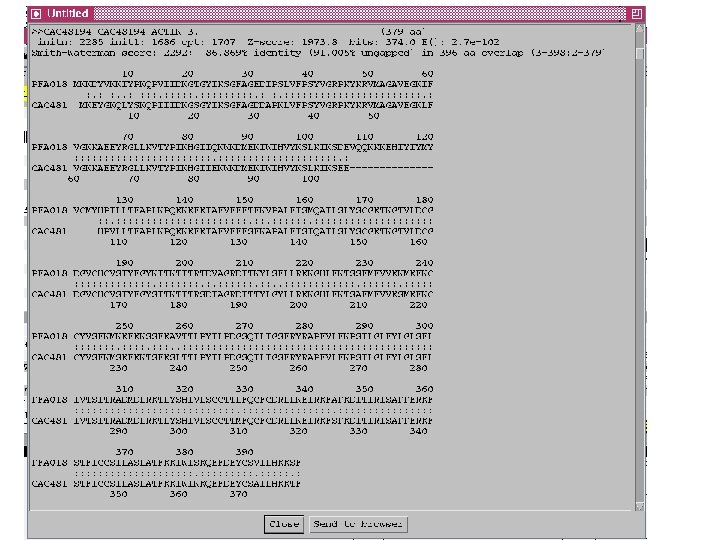
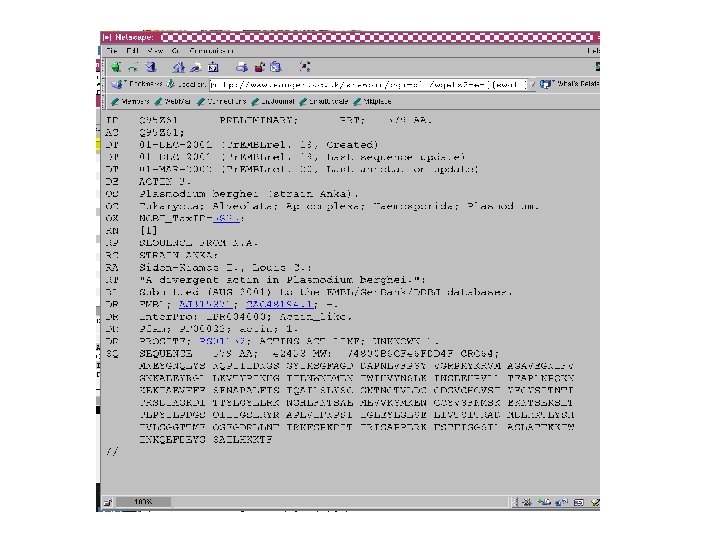
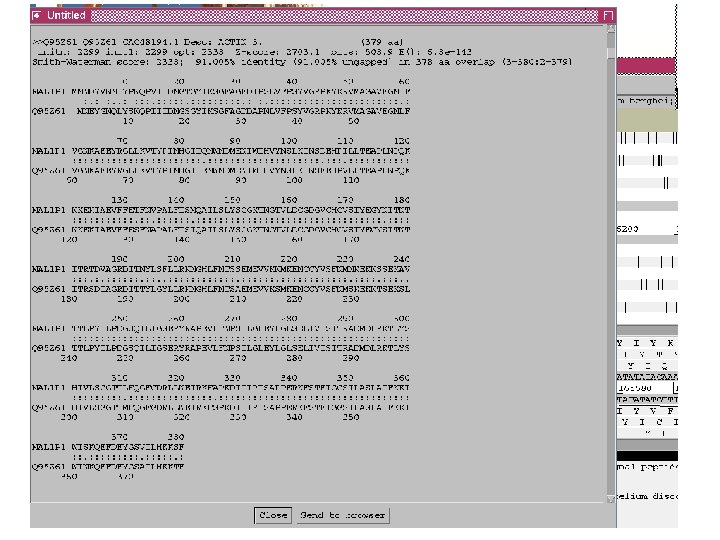
Curating gene models in Artemis Use of FASTA evidence






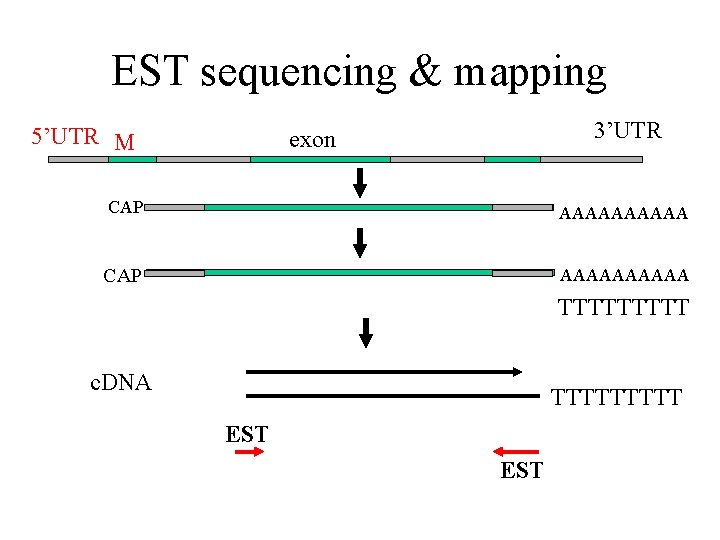
EST sequencing & mapping 5’UTR M intron exon stop 3’UTR CAP AAAAAAAAAA m. RNA TTTTT c. DNA TTTTT EST

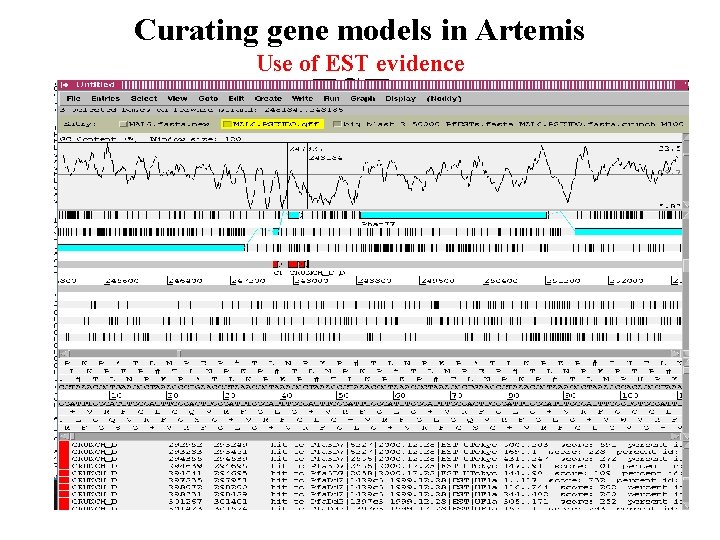
Curating gene models in Artemis Use of EST evidence ESTs

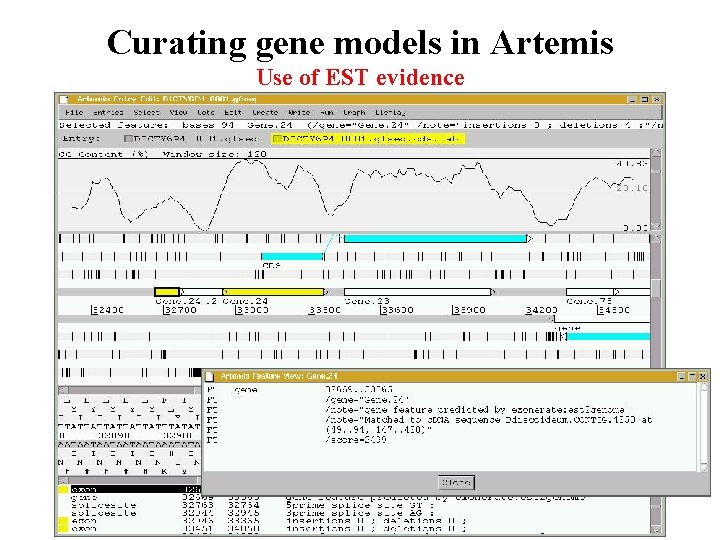
Curating gene models in Artemis Use of EST evidence

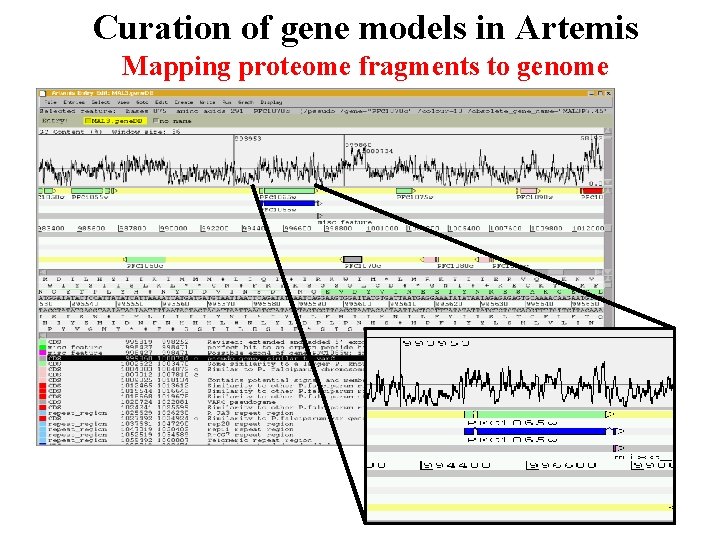
Curation of gene models in Artemis Mapping proteome fragments to genome

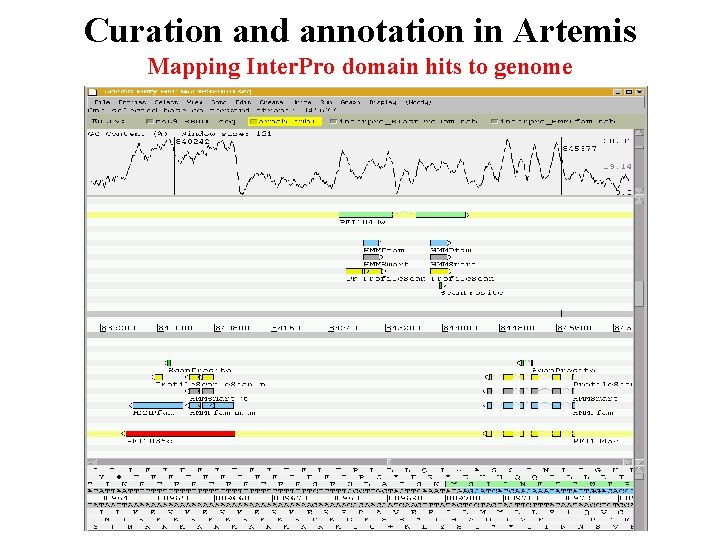
Curation and annotation in Artemis Mapping Inter. Pro domain hits to genome

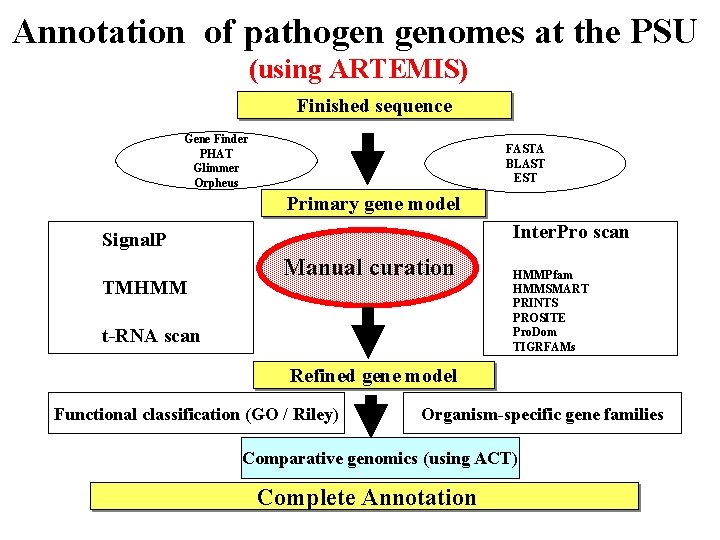
Annotation of pathogen genomes at the PSU (using ARTEMIS) Finished sequence Gene Finder PHAT Glimmer Orpheus FASTA BLAST EST Primary gene model Inter. Pro scan Signal. P TMHMM Manual curation t-RNA scan HMMPfam HMMSMART PRINTS PROSITE Pro. Dom TIGRFAMs Refined gene model Functional classification (GO / Riley) Organism-specific gene families Comparative genomics (using ACT) Complete Annotation

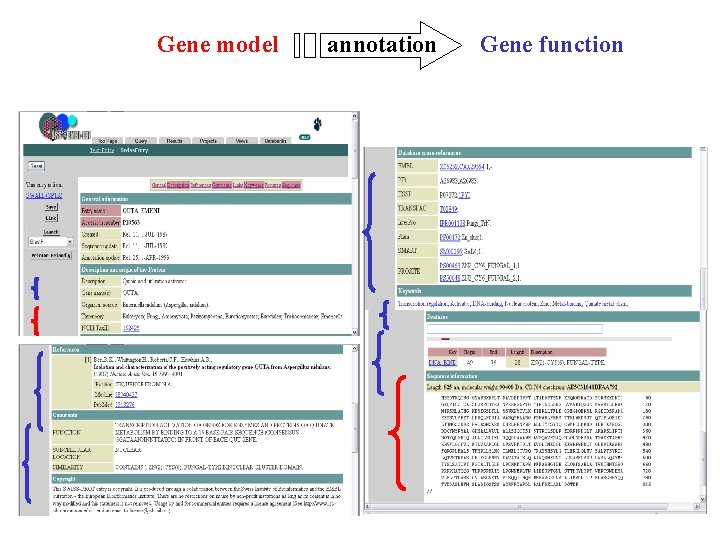
Gene model annotation Gene function

Top tips! Manual annotation. Use a several lines of evidence: - Run several available gene finding programs - Search programs: local (BLAST) and global (FASTA) alignments -Protein domains and motifs: Interpro (Pfam, prosite, SMART etc. ) -Transmembrane / signal peptide prediction (TMHMM, Signal. P) - Base your annotation on characterised proteins where possible (e. g. UNIPROT entry) - Read the literature (Pubmed entry)

Sanger Front page